Jiangwen Sun
Supervised Contrastive Vision Transformer for Breast Histopathological Image Classification
Apr 18, 2024Abstract:Invasive ductal carcinoma (IDC) is the most prevalent form of breast cancer. Breast tissue histopathological examination is critical in diagnosing and classifying breast cancer. Although existing methods have shown promising results, there is still room for improvement in the classification accuracy and generalization of IDC using histopathology images. We present a novel approach, Supervised Contrastive Vision Transformer (SupCon-ViT), for improving the classification of invasive ductal carcinoma in terms of accuracy and generalization by leveraging the inherent strengths and advantages of both transfer learning, i.e., pre-trained vision transformer, and supervised contrastive learning. Our results on a benchmark breast cancer dataset demonstrate that SupCon-Vit achieves state-of-the-art performance in IDC classification, with an F1-score of 0.8188, precision of 0.7692, and specificity of 0.8971, outperforming existing methods. In addition, the proposed model demonstrates resilience in scenarios with minimal labeled data, making it highly efficient in real-world clinical settings where labelled data is limited. Our findings suggest that supervised contrastive learning in conjunction with pre-trained vision transformers appears to be a viable strategy for an accurate classification of IDC, thus paving the way for a more efficient and reliable diagnosis of breast cancer through histopathological image analysis.
Highly Scalable Task Grouping for Deep Multi-Task Learning in Prediction of Epigenetic Events
Sep 24, 2022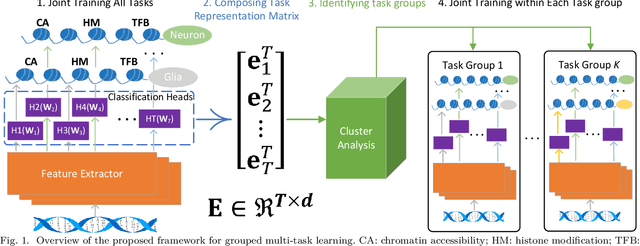
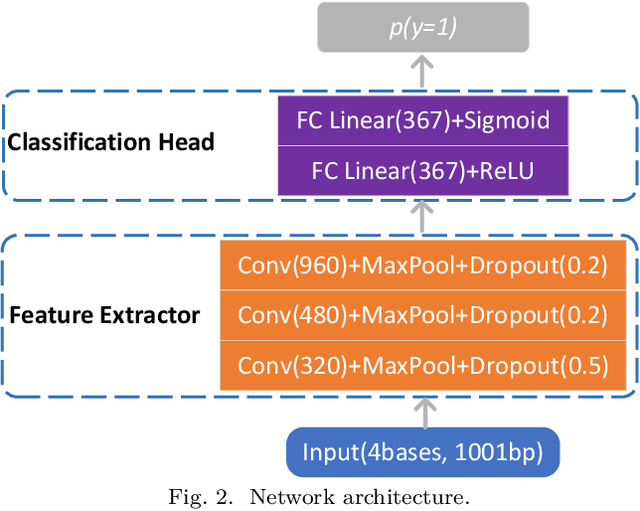
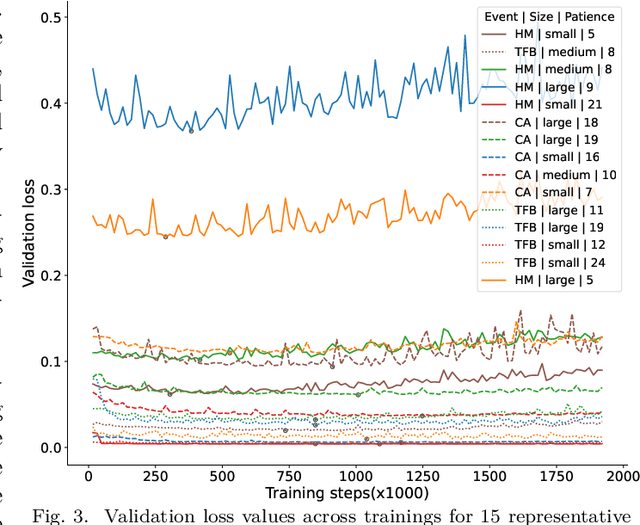
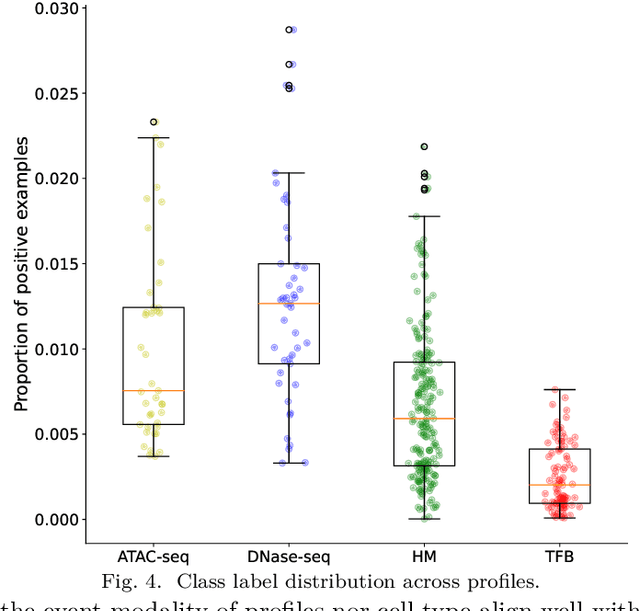
Abstract:Deep neural networks trained for predicting cellular events from DNA sequence have become emerging tools to help elucidate the biological mechanism underlying the associations identified in genome-wide association studies. To enhance the training, multi-task learning (MTL) has been commonly exploited in previous works where trained networks were needed for multiple profiles differing in either event modality or cell type. All existing works adopted a simple MTL framework where all tasks share a single feature extraction network. Such a strategy even though effective to certain extent leads to substantial negative transfer, meaning the existence of large portion of tasks for which models obtained through MTL perform worse than those by single task learning. There have been methods developed to address such negative transfer in other domains, such as computer vision. However, these methods are generally difficult to scale up to handle large amount of tasks. In this paper, we propose a highly scalable task grouping framework to address negative transfer by only jointly training tasks that are potentially beneficial to each other. The proposed method exploits the network weights associated with task specific classification heads that can be cheaply obtained by one-time joint training of all tasks. Our results using a dataset consisting of 367 epigenetic profiles demonstrate the effectiveness of the proposed approach and its superiority over baseline methods.
Edge Attention-based Multi-Relational Graph Convolutional Networks
May 20, 2018



Abstract:Graph convolutional network (GCN) is generalization of convolutional neural network (CNN) to work with arbitrarily structured graphs. A binary adjacency matrix is commonly used in training a GCN. Recently, the attention mechanism allows the network to learn a dynamic and adaptive aggregation of the neighborhood. We propose a new GCN model on the graphs where edges are characterized in multiple views or precisely in terms of multiple relationships. For instance, in chemical graph theory, compound structures are often represented by the hydrogen-depleted molecular graph where nodes correspond to atoms and edges correspond to chemical bonds. Multiple attributes can be important to characterize chemical bonds, such as atom pair (the types of atoms that a bond connects), aromaticity, and whether a bond is in a ring. The different attributes lead to different graph representations for the same molecule. There is growing interests in both chemistry and machine learning fields to directly learn molecular properties of compounds from the molecular graph, instead of from fingerprints predefined by chemists. The proposed GCN model, which we call edge attention-based multi-relational GCN (EAGCN), jointly learns attention weights and node features in graph convolution. For each bond attribute, a real-valued attention matrix is used to replace the binary adjacency matrix. By designing a dictionary for the edge attention, and forming the attention matrix of each molecule by looking up the dictionary, the EAGCN exploits correspondence between bonds in different molecules. The prediction of compound properties is based on the aggregated node features, which is independent of the varying molecule (graph) size. We demonstrate the efficacy of the EAGCN on multiple chemical datasets: Tox21, HIV, Freesolv, and Lipophilicity, and interpret the resultant attention weights.
VIGAN: Missing View Imputation with Generative Adversarial Networks
Nov 01, 2017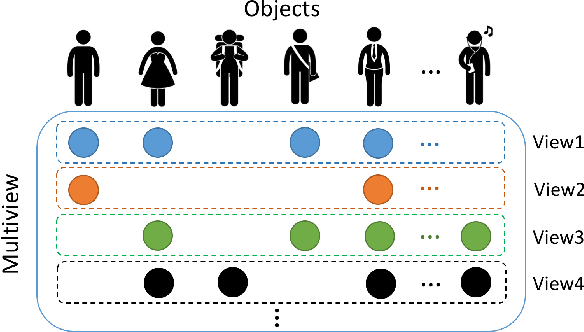
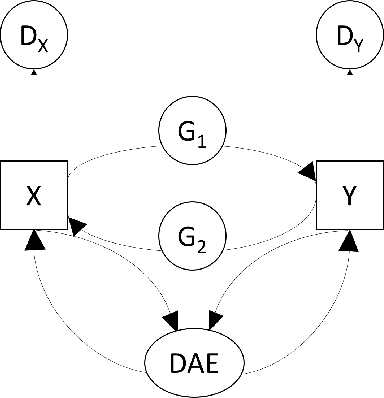
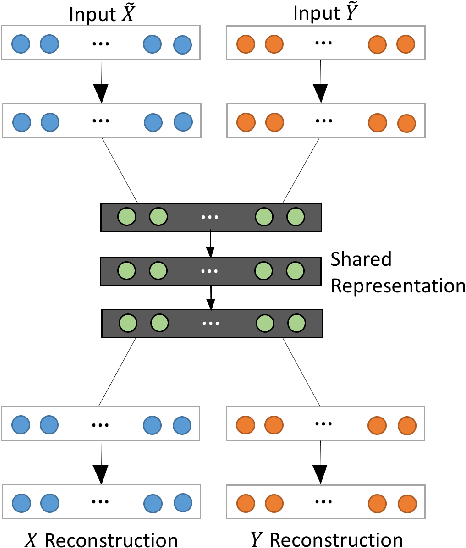
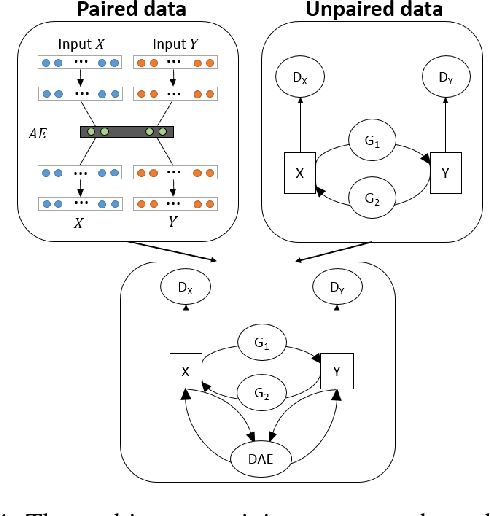
Abstract:In an era when big data are becoming the norm, there is less concern with the quantity but more with the quality and completeness of the data. In many disciplines, data are collected from heterogeneous sources, resulting in multi-view or multi-modal datasets. The missing data problem has been challenging to address in multi-view data analysis. Especially, when certain samples miss an entire view of data, it creates the missing view problem. Classic multiple imputations or matrix completion methods are hardly effective here when no information can be based on in the specific view to impute data for such samples. The commonly-used simple method of removing samples with a missing view can dramatically reduce sample size, thus diminishing the statistical power of a subsequent analysis. In this paper, we propose a novel approach for view imputation via generative adversarial networks (GANs), which we name by VIGAN. This approach first treats each view as a separate domain and identifies domain-to-domain mappings via a GAN using randomly-sampled data from each view, and then employs a multi-modal denoising autoencoder (DAE) to reconstruct the missing view from the GAN outputs based on paired data across the views. Then, by optimizing the GAN and DAE jointly, our model enables the knowledge integration for domain mappings and view correspondences to effectively recover the missing view. Empirical results on benchmark datasets validate the VIGAN approach by comparing against the state of the art. The evaluation of VIGAN in a genetic study of substance use disorders further proves the effectiveness and usability of this approach in life science.
On Multiplicative Multitask Feature Learning
Oct 24, 2016



Abstract:We investigate a general framework of multiplicative multitask feature learning which decomposes each task's model parameters into a multiplication of two components. One of the components is used across all tasks and the other component is task-specific. Several previous methods have been proposed as special cases of our framework. We study the theoretical properties of this framework when different regularization conditions are applied to the two decomposed components. We prove that this framework is mathematically equivalent to the widely used multitask feature learning methods that are based on a joint regularization of all model parameters, but with a more general form of regularizers. Further, an analytical formula is derived for the across-task component as related to the task-specific component for all these regularizers, leading to a better understanding of the shrinkage effect. Study of this framework motivates new multitask learning algorithms. We propose two new learning formulations by varying the parameters in the proposed framework. Empirical studies have revealed the relative advantages of the two new formulations by comparing with the state of the art, which provides instructive insights into the feature learning problem with multiple tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge