Ivan Viola
Augmenting a Large Language Model with a Combination of Text and Visual Data for Conversational Visualization of Global Geospatial Data
Jan 16, 2025

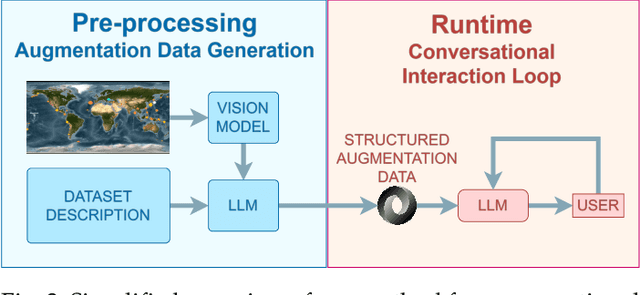

Abstract:We present a method for augmenting a Large Language Model (LLM) with a combination of text and visual data to enable accurate question answering in visualization of scientific data, making conversational visualization possible. LLMs struggle with tasks like visual data interaction, as they lack contextual visual information. We address this problem by merging a text description of a visualization and dataset with snapshots of the visualization. We extract their essential features into a structured text file, highly compact, yet descriptive enough to appropriately augment the LLM with contextual information, without any fine-tuning. This approach can be applied to any visualization that is already finally rendered, as long as it is associated with some textual description.
Dr. KID: Direct Remeshing and K-set Isometric Decomposition for Scalable Physicalization of Organic Shapes
Apr 06, 2023



Abstract:Dr. KID is an algorithm that uses isometric decomposition for the physicalization of potato-shaped organic models in a puzzle fashion. The algorithm begins with creating a simple, regular triangular surface mesh of organic shapes, followed by iterative k-means clustering and remeshing. For clustering, we need similarity between triangles (segments) which is defined as a distance function. The distance function maps each triangle's shape to a single point in the virtual 3D space. Thus, the distance between the triangles indicates their degree of dissimilarity. K-means clustering uses this distance and sorts of segments into k classes. After this, remeshing is applied to minimize the distance between triangles within the same cluster by making their shapes identical. Clustering and remeshing are repeated until the distance between triangles in the same cluster reaches an acceptable threshold. We adopt a curvature-aware strategy to determine the surface thickness and finalize puzzle pieces for 3D printing. Identical hinges and holes are created for assembling the puzzle components. For smoother outcomes, we use triangle subdivision along with curvature-aware clustering, generating curved triangular patches for 3D printing. Our algorithm was evaluated using various models, and the 3D-printed results were analyzed. Findings indicate that our algorithm performs reliably on target organic shapes with minimal loss of input geometry.
Differentiable Electron Microscopy Simulation: Methods and Applications for Visualization
May 08, 2022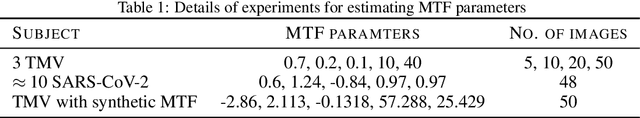
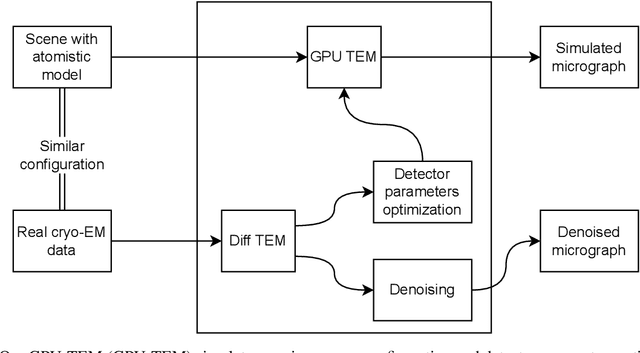
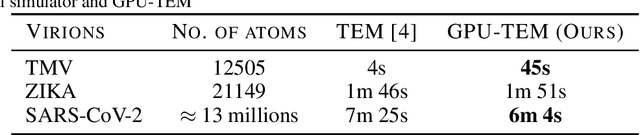
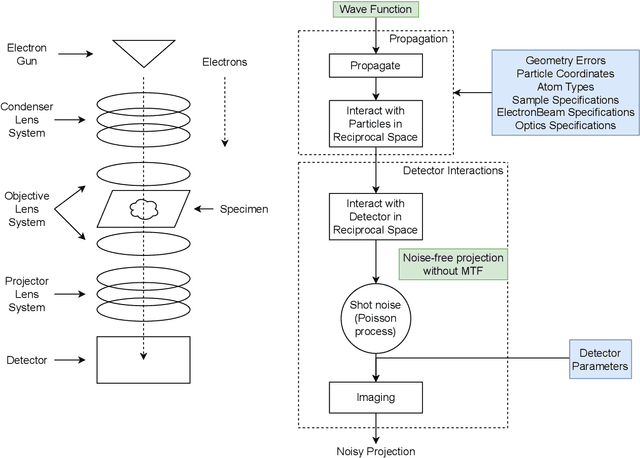
Abstract:We propose a new microscopy simulation system that can depict atomistic models in a micrograph visual style, similar to results of physical electron microscopy imaging. This system is scalable, able to represent simulation of electron microscopy of tens of viral particles and synthesizes the image faster than previous methods. On top of that, the simulator is differentiable, both its deterministic as well as stochastic stages that form signal and noise representations in the micrograph. This notable property has the capability for solving inverse problems by means of optimization and thus allows for generation of microscopy simulations using the parameter settings estimated from real data. We demonstrate this learning capability through two applications: (1) estimating the parameters of the modulation transfer function defining the detector properties of the simulated and real micrographs, and (2) denoising the real data based on parameters trained from the simulated examples. While current simulators do not support any parameter estimation due to their forward design, we show that the results obtained using estimated parameters are very similar to the results of real micrographs. Additionally, we evaluate the denoising capabilities of our approach and show that the results showed an improvement over state-of-the-art methods. Denoised micrographs exhibit less noise in the tilt-series tomography reconstructions, ultimately reducing the visual dominance of noise in direct volume rendering of microscopy tomograms.
The Ultrasound Visualization Pipeline - A Survey
Jun 18, 2012



Abstract:Ultrasound is one of the most frequently used imaging modality in medicine. The high spatial resolution, its interactive nature and non-invasiveness makes it the first choice in many examinations. Image interpretation is one of ultrasound's main challenges. Much training is required to obtain a confident skill level in ultrasound-based diagnostics. State-of-the-art graphics techniques is needed to provide meaningful visualizations of ultrasound in real-time. In this paper we present the process-pipeline for ultrasound visualization, including an overview of the tasks performed in the specific steps. To provide an insight into the trends of ultrasound visualization research, we have selected a set of significant publications and divided them into a technique-based taxonomy covering the topics pre-processing, segmentation, registration, rendering and augmented reality. For the different technique types we discuss the difference between ultrasound-based techniques and techniques for other modalities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge