Ngan Nguyen
CTPD: Cross Tokenizer Preference Distillation
Jan 17, 2026Abstract:While knowledge distillation has seen widespread use in pre-training and instruction tuning, its application to aligning language models with human preferences remains underexplored, particularly in the more realistic cross-tokenizer setting. The incompatibility of tokenization schemes between teacher and student models has largely prevented fine-grained, white-box distillation of preference information. To address this gap, we propose Cross-Tokenizer Preference Distillation (CTPD), the first unified framework for transferring human-aligned behavior between models with heterogeneous tokenizers. CTPD introduces three key innovations: (1) Aligned Span Projection, which maps teacher and student tokens to shared character-level spans for precise supervision transfer; (2) a cross-tokenizer adaptation of Token-level Importance Sampling (TIS-DPO) for improved credit assignment; and (3) a Teacher-Anchored Reference, allowing the student to directly leverage the teacher's preferences in a DPO-style objective. Our theoretical analysis grounds CTPD in importance sampling, and experiments across multiple benchmarks confirm its effectiveness, with significant performance gains over existing methods. These results establish CTPD as a practical and general solution for preference distillation across diverse tokenization schemes, opening the door to more accessible and efficient alignment of language models.
"Some other poor soul's problems": a peer recommendation intervention for health-related social support
Sep 12, 2022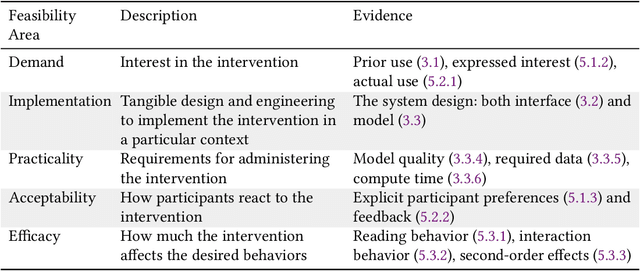
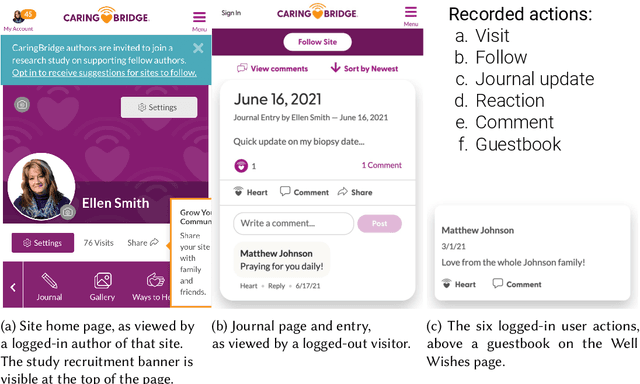
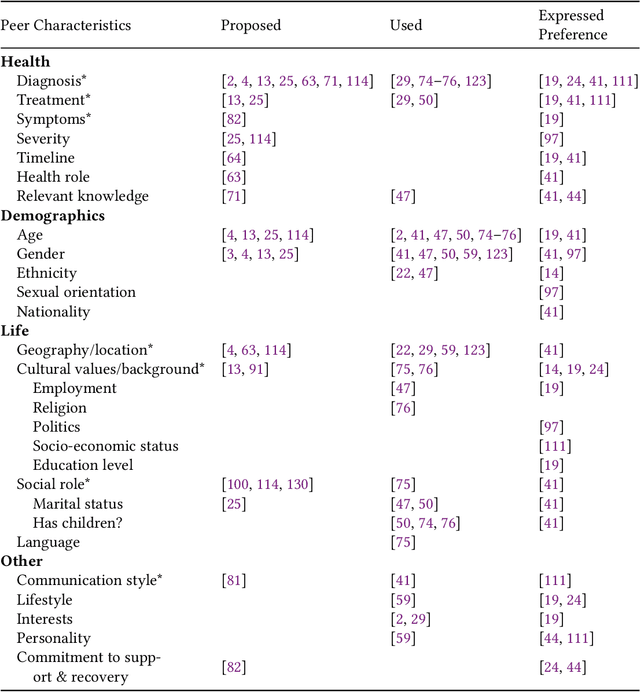
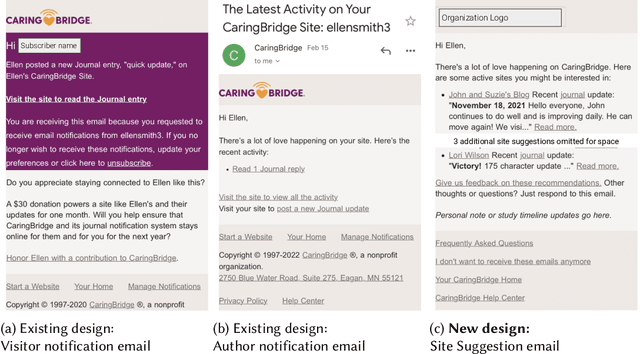
Abstract:Online health communities (OHCs) offer the promise of connecting with supportive peers. Forming these connections first requires finding relevant peers - a process that can be time-consuming. Peer recommendation systems are a computational approach to make finding peers easier during a health journey. By encouraging OHC users to alter their online social networks, peer recommendations could increase available support. But these benefits are hypothetical and based on mixed, observational evidence. To experimentally evaluate the effect of peer recommendations, we conceptualize these systems as health interventions designed to increase specific beneficial connection behaviors. In this paper, we designed a peer recommendation intervention to increase two behaviors: reading about peer experiences and interacting with peers. We conducted an initial feasibility assessment of this intervention by conducting a 12-week field study in which 79 users of CaringBridge received weekly peer recommendations via email. Our results support the usefulness and demand for peer recommendation and suggest benefits to evaluating larger peer recommendation interventions. Our contributions include practical guidance on the development and evaluation of peer recommendation interventions for OHCs.
Differentiable Electron Microscopy Simulation: Methods and Applications for Visualization
May 08, 2022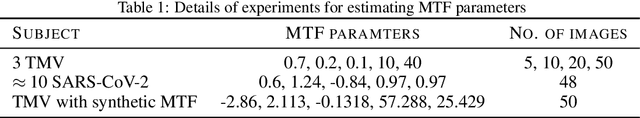
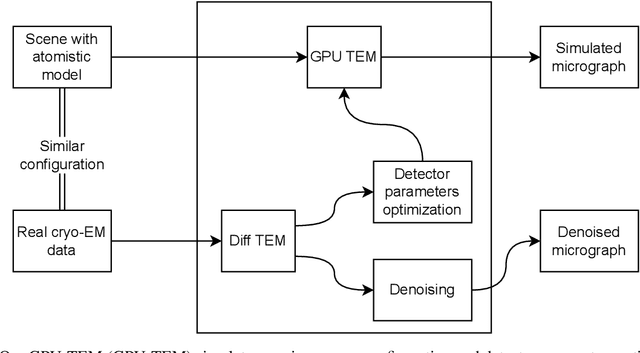
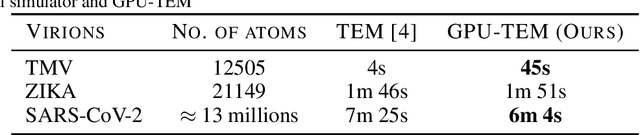
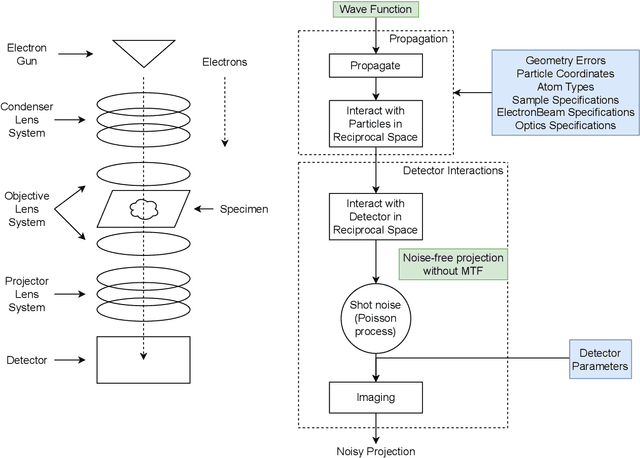
Abstract:We propose a new microscopy simulation system that can depict atomistic models in a micrograph visual style, similar to results of physical electron microscopy imaging. This system is scalable, able to represent simulation of electron microscopy of tens of viral particles and synthesizes the image faster than previous methods. On top of that, the simulator is differentiable, both its deterministic as well as stochastic stages that form signal and noise representations in the micrograph. This notable property has the capability for solving inverse problems by means of optimization and thus allows for generation of microscopy simulations using the parameter settings estimated from real data. We demonstrate this learning capability through two applications: (1) estimating the parameters of the modulation transfer function defining the detector properties of the simulated and real micrographs, and (2) denoising the real data based on parameters trained from the simulated examples. While current simulators do not support any parameter estimation due to their forward design, we show that the results obtained using estimated parameters are very similar to the results of real micrographs. Additionally, we evaluate the denoising capabilities of our approach and show that the results showed an improvement over state-of-the-art methods. Denoised micrographs exhibit less noise in the tilt-series tomography reconstructions, ultimately reducing the visual dominance of noise in direct volume rendering of microscopy tomograms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge