Isabelle Salmon
Exploring Foundation Models Fine-Tuning for Cytology Classification
Nov 22, 2024
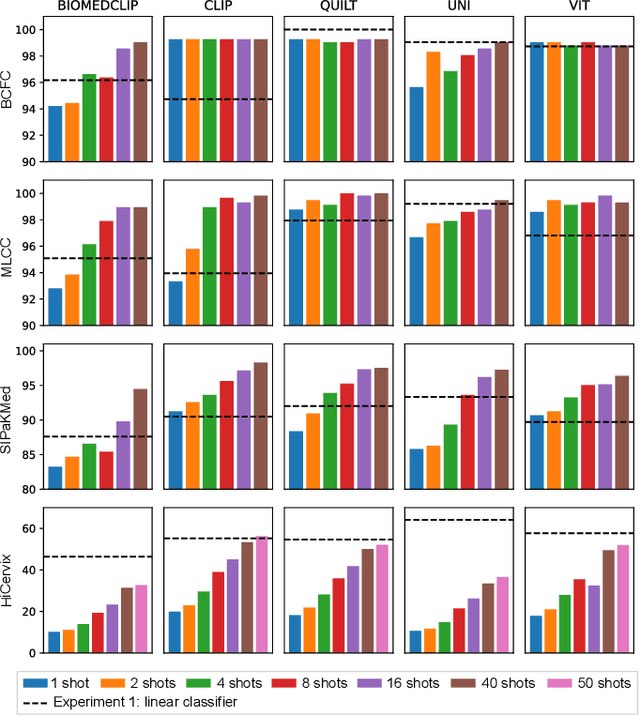

Abstract:Cytology slides are essential tools in diagnosing and staging cancer, but their analysis is time-consuming and costly. Foundation models have shown great potential to assist in these tasks. In this paper, we explore how existing foundation models can be applied to cytological classification. More particularly, we focus on low-rank adaptation, a parameter-efficient fine-tuning method suited to few-shot learning. We evaluated five foundation models across four cytological classification datasets. Our results demonstrate that fine-tuning the pre-trained backbones with LoRA significantly improves model performance compared to fine-tuning only the classifier head, achieving state-of-the-art results on both simple and complex classification tasks while requiring fewer data samples.
Interpretable HER2 scoring by evaluating clinical Guidelines through a weakly supervised, constrained Deep Learning Approach
Nov 17, 2022



Abstract:The evaluation of the Human Epidermal growth factor Receptor-2 (HER2) expression is an important prognostic biomarker for breast cancer treatment selection. However, HER2 scoring has notoriously high interobserver variability due to stain variations between centers and the need to estimate visually the staining intensity in specific percentages of tumor area. In this paper, focusing on the interpretability of HER2 scoring by a pathologist, we propose a semi-automatic, two-stage deep learning approach that directly evaluates the clinical HER2 guidelines defined by the American Society of Clinical Oncology/ College of American Pathologists (ASCO/CAP). In the first stage, we segment the invasive tumor over the user-indicated Region of Interest (ROI). Then, in the second stage, we classify the tumor tissue into four HER2 classes. For the classification stage, we use weakly supervised, constrained optimization to find a model that classifies cancerous patches such that the tumor surface percentage meets the guidelines specification of each HER2 class. We end the second stage by freezing the model and refining its output logits in a supervised way to all slide labels in the training set. To ensure the quality of our dataset's labels, we conducted a multi-pathologist HER2 scoring consensus. For the assessment of doubtful cases where no consensus was found, our model can help by interpreting its HER2 class percentages output. We achieve a performance of 0.78 in F1-score on the test set while keeping our model interpretable for the pathologist, hopefully contributing to interpretable AI models in digital pathology.
Initial condition assessment for reaction-diffusion glioma growth models: A translational MRI/histology (in)validation study
Feb 02, 2021



Abstract:Diffuse gliomas are highly infiltrative tumors whose early diagnosis and follow-up usually rely on magnetic resonance imaging (MRI). However, the limited sensitivity of this technique makes it impossible to directly assess the extent of the glioma cell invasion, leading to sub-optimal treatment planing. Reaction-diffusion growth models have been proposed for decades to extrapolate glioma cell infiltration beyond margins visible on MRI and predict its spatial-temporal evolution. These models nevertheless require an initial condition, that is the tumor cell density values at every location of the brain at diagnosis time. Several works have proposed to relate the tumor cell density function to abnormality outlines visible on MRI but the underlying assumptions have never been verified so far. In this work we propose to verify these assumptions by stereotactic histological analysis of a non-operated brain with glioblastoma using a tailored 3D-printed slicer. Cell density maps are computed from histological slides using a deep learning approach. The density maps are then registered to a postmortem MR image and related to an MR-derived geodesic distance map to the tumor core. The relation between the edema outlines visible on T2 FLAIR MRI and the distance to the core is also investigated. Our results suggest that (i) the previously suggested exponential decrease of the tumor cell density with the distance to the tumor core is not unreasonable but (ii) the edema outlines may in general not correspond to a cell density iso-contour and (iii) the commonly adopted tumor cell density value at these outlines is likely overestimated. These findings highlight the limitations of using conventional MRI to derive glioma cell density maps and point out the need of validating other methods to initialize reaction-diffusion growth models and make them usable in clinical practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge