Exploring Foundation Models Fine-Tuning for Cytology Classification
Paper and Code
Nov 22, 2024
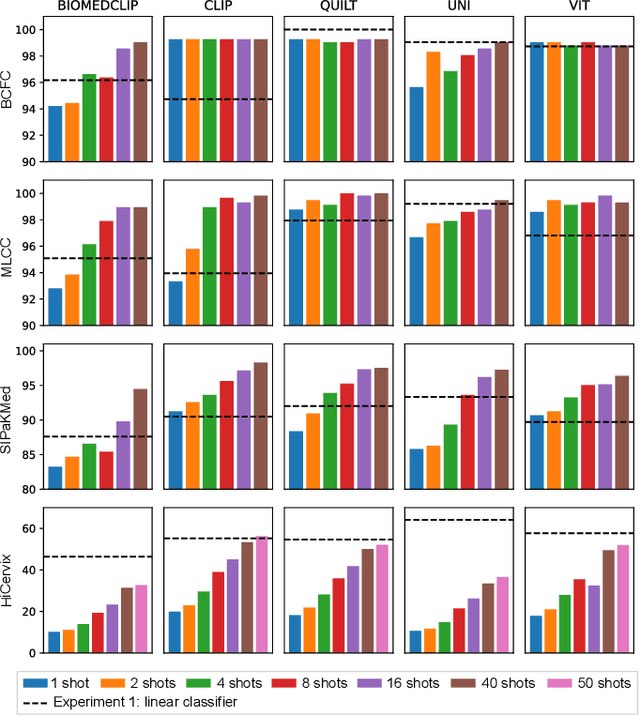

Cytology slides are essential tools in diagnosing and staging cancer, but their analysis is time-consuming and costly. Foundation models have shown great potential to assist in these tasks. In this paper, we explore how existing foundation models can be applied to cytological classification. More particularly, we focus on low-rank adaptation, a parameter-efficient fine-tuning method suited to few-shot learning. We evaluated five foundation models across four cytological classification datasets. Our results demonstrate that fine-tuning the pre-trained backbones with LoRA significantly improves model performance compared to fine-tuning only the classifier head, achieving state-of-the-art results on both simple and complex classification tasks while requiring fewer data samples.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge