Hugo Mark Horlings
Training state-of-the-art pathology foundation models with orders of magnitude less data
Apr 07, 2025Abstract:The field of computational pathology has recently seen rapid advances driven by the development of modern vision foundation models (FMs), typically trained on vast collections of pathology images. Recent studies demonstrate that increasing the training data set and model size and integrating domain-specific image processing techniques can significantly enhance the model's performance on downstream tasks. Building on these insights, our work incorporates several recent modifications to the standard DINOv2 framework from the literature to optimize the training of pathology FMs. We also apply a post-training procedure for fine-tuning models on higher-resolution images to further enrich the information encoded in the embeddings. We present three novel pathology FMs trained on up to two orders of magnitude fewer WSIs than those used to train other state-of-the-art FMs while demonstrating a comparable or superior performance on downstream tasks. Even the model trained on TCGA alone (12k WSIs) outperforms most existing FMs and, on average, matches Virchow2, the second-best FM published to date. This suggests that there still remains a significant potential for further improving the models and algorithms used to train pathology FMs to take full advantage of the vast data collections.
PatchSorter: A High Throughput Deep Learning Digital Pathology Tool for Object Labeling
Jul 13, 2023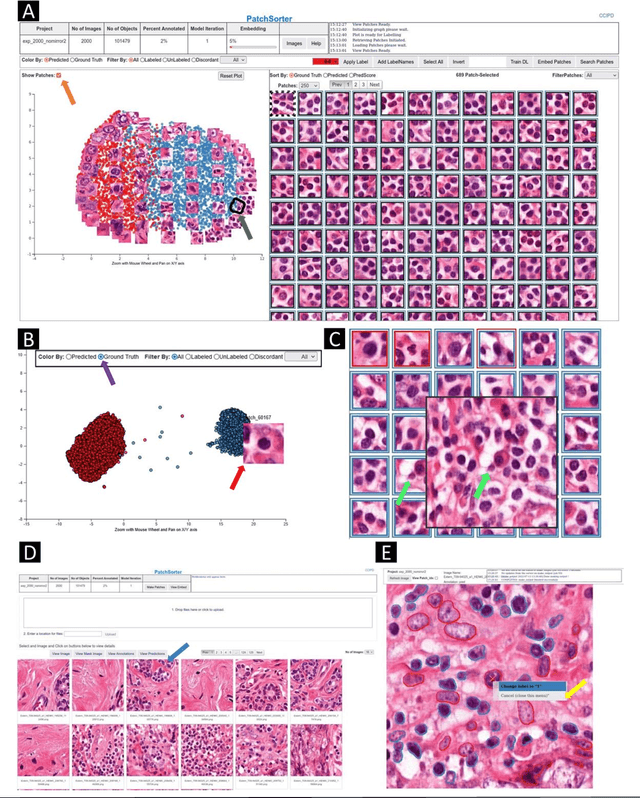
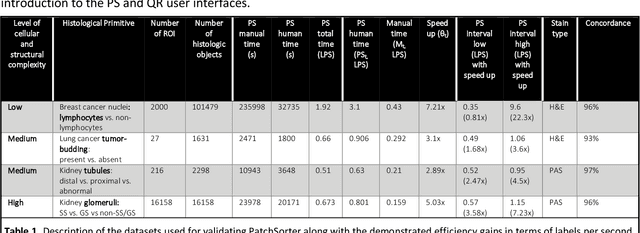
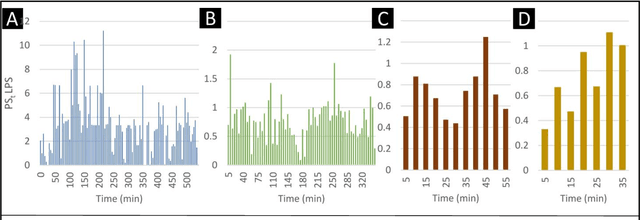
Abstract:The discovery of patterns associated with diagnosis, prognosis, and therapy response in digital pathology images often requires intractable labeling of large quantities of histological objects. Here we release an open-source labeling tool, PatchSorter, which integrates deep learning with an intuitive web interface. Using >100,000 objects, we demonstrate a >7x improvement in labels per second over unaided labeling, with minimal impact on labeling accuracy, thus enabling high-throughput labeling of large datasets.
WeakSTIL: Weak whole-slide image level stromal tumor infiltrating lymphocyte scores are all you need
Sep 13, 2021
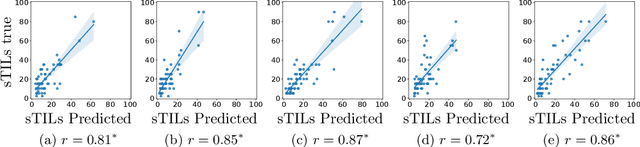
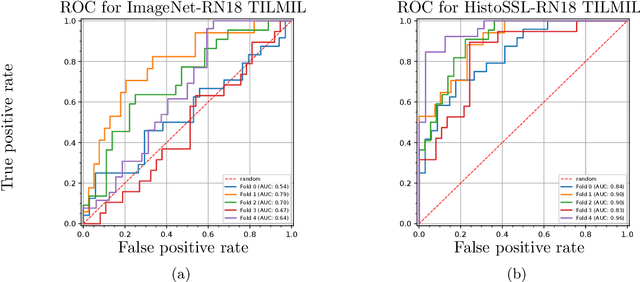
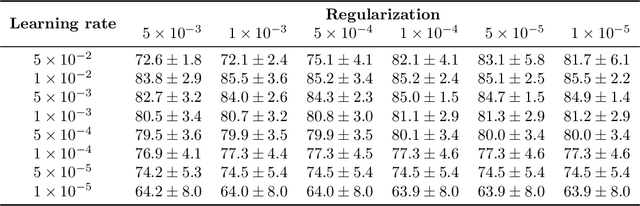
Abstract:We present WeakSTIL, an interpretable two-stage weak label deep learning pipeline for scoring the percentage of stromal tumor infiltrating lymphocytes (sTIL%) in H&E-stained whole-slide images (WSIs) of breast cancer tissue. The sTIL% score is a prognostic and predictive biomarker for many solid tumor types. However, due to the high labeling efforts and high intra- and interobserver variability within and between expert annotators, this biomarker is currently not used in routine clinical decision making. WeakSTIL compresses tiles of a WSI using a feature extractor pre-trained with self-supervised learning on unlabeled histopathology data and learns to predict precise sTIL% scores for each tile in the tumor bed by using a multiple instance learning regressor that only requires a weak WSI-level label. By requiring only a weak label, we overcome the large annotation efforts required to train currently existing TIL detection methods. We show that WeakSTIL is at least as good as other TIL detection methods when predicting the WSI-level sTIL% score, reaching a coefficient of determination of $0.45\pm0.15$ when compared to scores generated by an expert pathologist, and an AUC of $0.89\pm0.05$ when treating it as the clinically interesting sTIL-high vs sTIL-low classification task. Additionally, we show that the intermediate tile-level predictions of WeakSTIL are highly interpretable, which suggests that WeakSTIL pays attention to latent features related to the number of TILs and the tissue type. In the future, WeakSTIL may be used to provide consistent and interpretable sTIL% predictions to stratify breast cancer patients into targeted therapy arms.
DeepSMILE: Self-supervised heterogeneity-aware multiple instance learning for DNA damage response defect classification directly from H&E whole-slide images
Jul 28, 2021

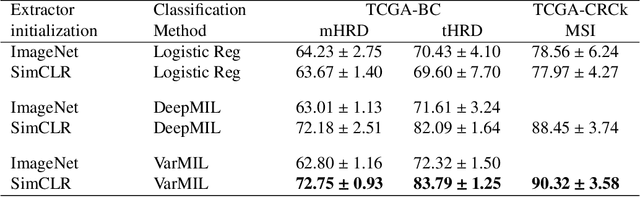
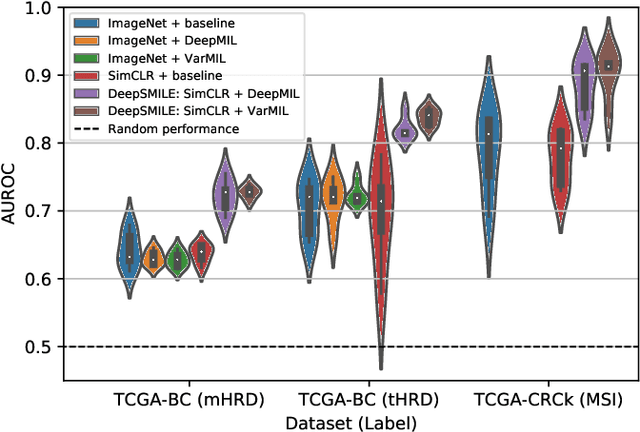
Abstract:We propose a Deep learning-based weak label learning method for analysing whole slide images (WSIs) of Hematoxylin and Eosin (H&E) stained tumorcells not requiring pixel-level or tile-level annotations using Self-supervised pre-training and heterogeneity-aware deep Multiple Instance LEarning (DeepSMILE). We apply DeepSMILE to the task of Homologous recombination deficiency (HRD) and microsatellite instability (MSI) prediction. We utilize contrastive self-supervised learning to pre-train a feature extractor on histopathology tiles of cancer tissue. Additionally, we use variability-aware deep multiple instance learning to learn the tile feature aggregation function while modeling tumor heterogeneity. Compared to state-of-the-art genomic label classification methods, DeepSMILE improves classification performance for HRD from $70.43\pm4.10\%$ to $83.79\pm1.25\%$ AUC and MSI from $78.56\pm6.24\%$ to $90.32\pm3.58\%$ AUC in a multi-center breast and colorectal cancer dataset, respectively. These improvements suggest we can improve genomic label classification performance without collecting larger datasets. In the future, this may reduce the need for expensive genome sequencing techniques, provide personalized therapy recommendations based on widely available WSIs of cancer tissue, and improve patient care with quicker treatment decisions - also in medical centers without access to genome sequencing resources.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge