Andreas Panteli
WeakSTIL: Weak whole-slide image level stromal tumor infiltrating lymphocyte scores are all you need
Sep 13, 2021
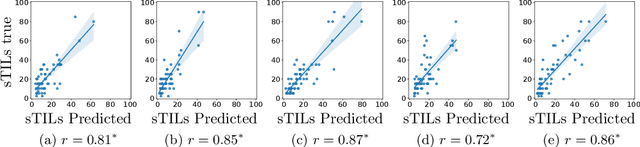
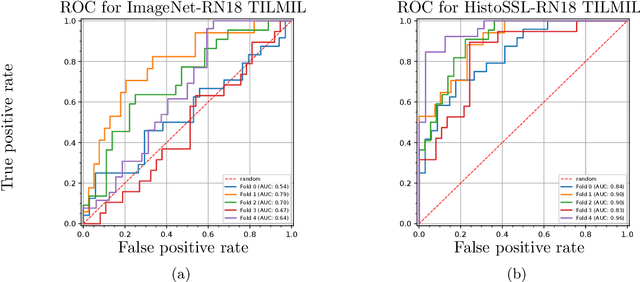
Abstract:We present WeakSTIL, an interpretable two-stage weak label deep learning pipeline for scoring the percentage of stromal tumor infiltrating lymphocytes (sTIL%) in H&E-stained whole-slide images (WSIs) of breast cancer tissue. The sTIL% score is a prognostic and predictive biomarker for many solid tumor types. However, due to the high labeling efforts and high intra- and interobserver variability within and between expert annotators, this biomarker is currently not used in routine clinical decision making. WeakSTIL compresses tiles of a WSI using a feature extractor pre-trained with self-supervised learning on unlabeled histopathology data and learns to predict precise sTIL% scores for each tile in the tumor bed by using a multiple instance learning regressor that only requires a weak WSI-level label. By requiring only a weak label, we overcome the large annotation efforts required to train currently existing TIL detection methods. We show that WeakSTIL is at least as good as other TIL detection methods when predicting the WSI-level sTIL% score, reaching a coefficient of determination of $0.45\pm0.15$ when compared to scores generated by an expert pathologist, and an AUC of $0.89\pm0.05$ when treating it as the clinically interesting sTIL-high vs sTIL-low classification task. Additionally, we show that the intermediate tile-level predictions of WeakSTIL are highly interpretable, which suggests that WeakSTIL pays attention to latent features related to the number of TILs and the tissue type. In the future, WeakSTIL may be used to provide consistent and interpretable sTIL% predictions to stratify breast cancer patients into targeted therapy arms.
Sparse-Shot Learning for Extremely Many Localisations
Apr 21, 2021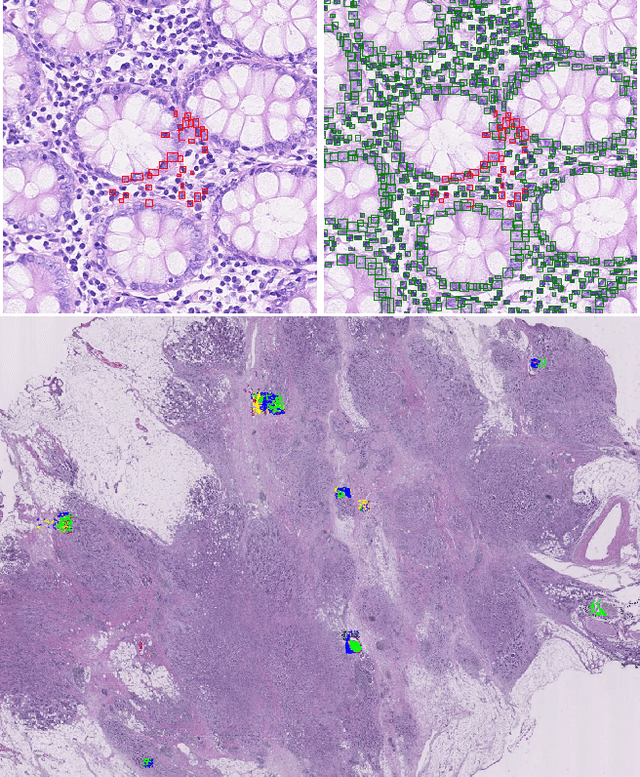
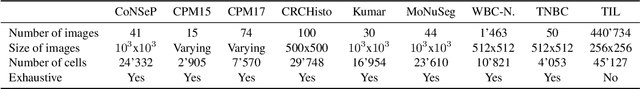
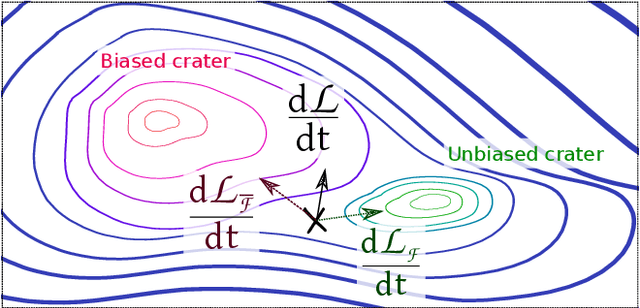
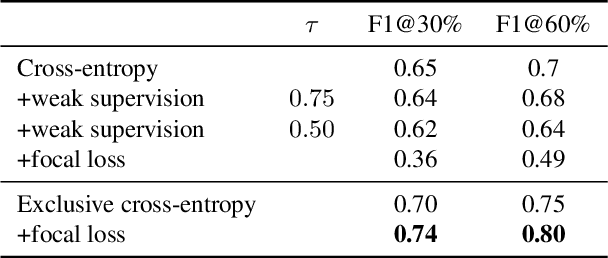
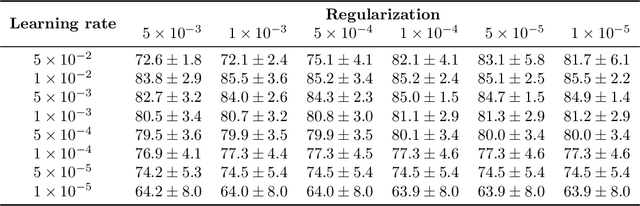
Abstract:Object localisation is typically considered in the context of regular images, for instance depicting objects like people or cars. In these images there is typically a relatively small number of instances per image per class, which usually is manageable to annotate. However, outside the realm of regular images we are often confronted with a different situation. In computational pathology digitised tissue sections are extremely large images, whose dimensions quickly exceed 250'000x250'000 pixels, where relevant objects, such as tumour cells or lymphocytes can quickly number in the millions. Annotating them all is practically impossible and annotating sparsely a few, out of many more, is the only possibility. Unfortunately, learning from sparse annotations, or sparse-shot learning, clashes with standard supervised learning because what is not annotated is treated as a negative. However, assigning negative labels to what are true positives leads to confusion in the gradients and biased learning. To this end, we present exclusive cross entropy, which slows down the biased learning by examining the second-order loss derivatives in order to drop the loss terms corresponding to likely biased terms. Experiments on nine datasets and two different localisation tasks, detection with YOLLO and segmentation with Unet, show that we obtain considerable improvements compared to cross entropy or focal loss, while often reaching the best possible performance for the model with only 10-40 of annotations.
Tracking-Assisted Segmentation of Biological Cells
Oct 19, 2019


Abstract:U-Net and its variants have been demonstrated to work sufficiently well in biological cell tracking and segmentation. However, these methods still suffer in the presence of complex processes such as collision of cells, mitosis and apoptosis. In this paper, we augment U-Net with Siamese matching-based tracking and propose to track individual nuclei over time. By modelling the behavioural pattern of the cells, we achieve improved segmentation and tracking performances through a re-segmentation procedure. Our preliminary investigations on the Fluo-N2DH-SIM+ and Fluo-N2DH-GOWT1 datasets demonstrate that absolute improvements of up to 3.8 % and 3.4% can be obtained in segmentation and tracking accuracy, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge