Heidi Seibold
What Makes Forest-Based Heterogeneous Treatment Effect Estimators Work?
Jun 21, 2022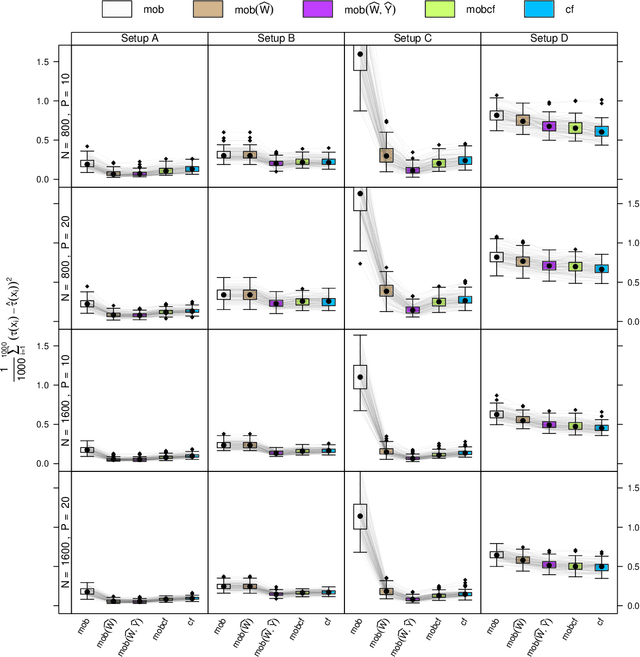
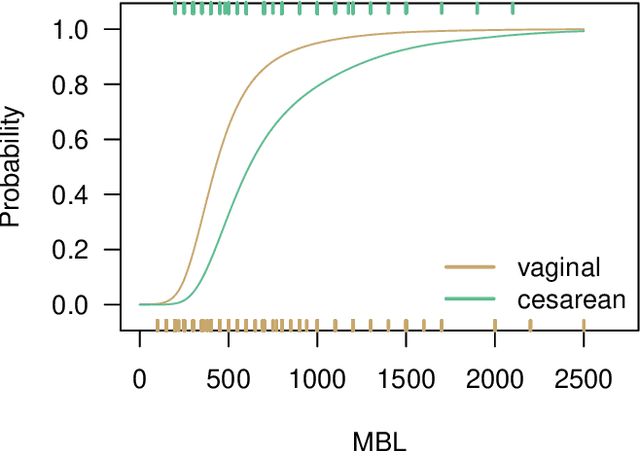
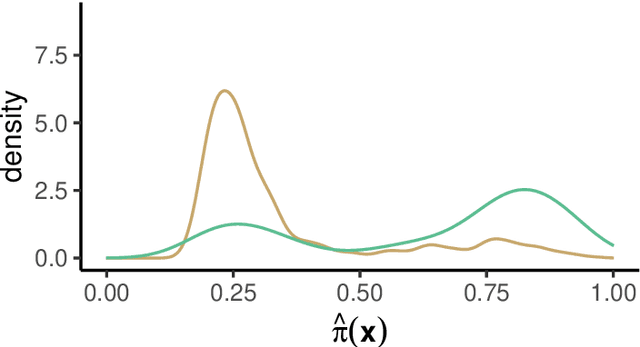
Abstract:Estimation of heterogeneous treatment effects (HTE) is of prime importance in many disciplines, ranging from personalized medicine to economics among many others. Random forests have been shown to be a flexible and powerful approach to HTE estimation in both randomized trials and observational studies. In particular "causal forests", introduced by Athey, Tibshirani and Wager (2019), along with the R implementation in package grf were rapidly adopted. A related approach, called "model-based forests", that is geared towards randomized trials and simultaneously captures effects of both prognostic and predictive variables, was introduced by Seibold, Zeileis and Hothorn (2018) along with a modular implementation in the R package model4you. Here, we present a unifying view that goes beyond the theoretical motivations and investigates which computational elements make causal forests so successful and how these can be blended with the strengths of model-based forests. To do so, we show that both methods can be understood in terms of the same parameters and model assumptions for an additive model under L2 loss. This theoretical insight allows us to implement several flavors of "model-based causal forests" and dissect their different elements in silico. The original causal forests and model-based forests are compared with the new blended versions in a benchmark study exploring both randomized trials and observational settings. In the randomized setting, both approaches performed akin. If confounding was present in the data generating process, we found local centering of the treatment indicator with the corresponding propensities to be the main driver for good performance. Local centering of the outcome was less important, and might be replaced or enhanced by simultaneous split selection with respect to both prognostic and predictive effects.
Survival Forests under Test: Impact of the Proportional Hazards Assumption on Prognostic and Predictive Forests for ALS Survival
Feb 05, 2019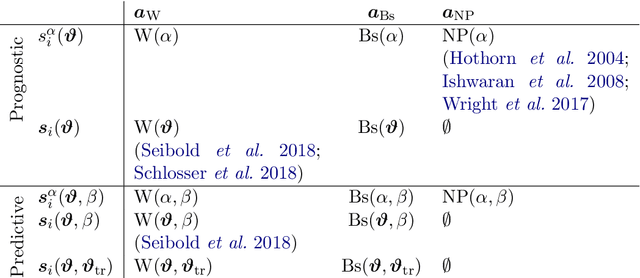
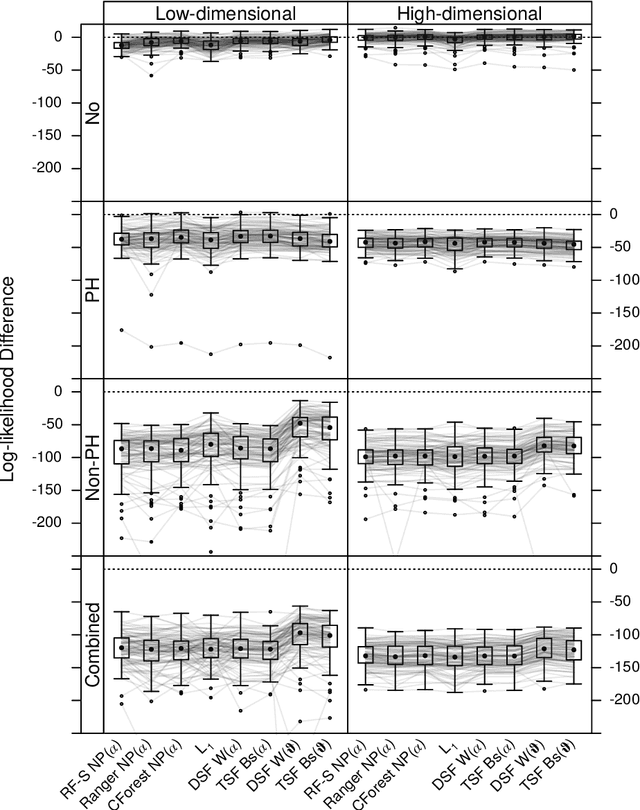
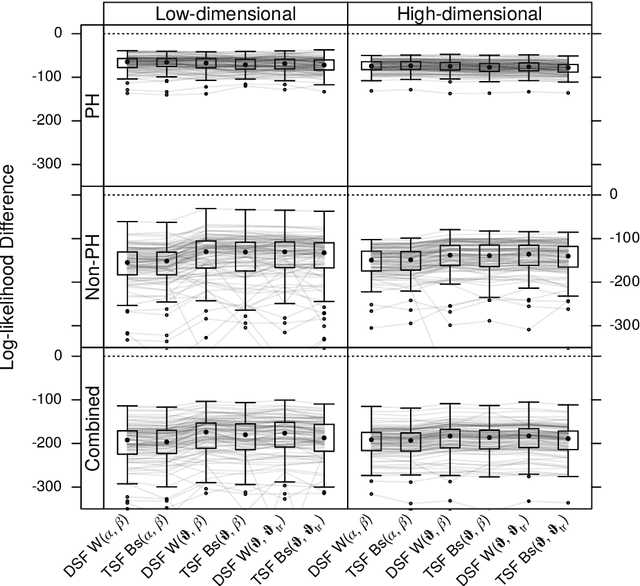
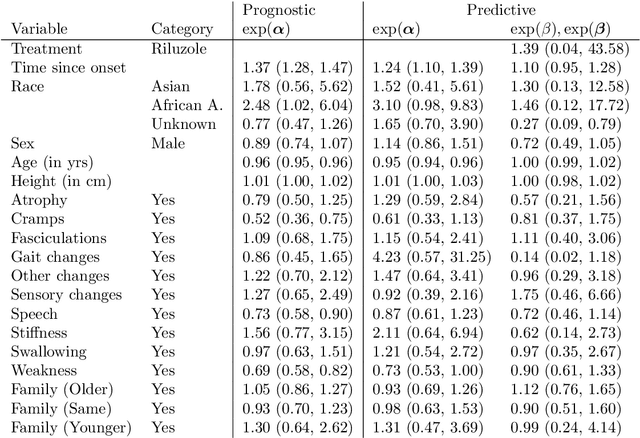
Abstract:We investigate the effect of the proportional hazards assumption on prognostic and predictive models of the survival time of patients suffering from amyotrophic lateral sclerosis (ALS). We theoretically compare the underlying model formulations of several variants of survival forests and implementations thereof, including random forests for survival, conditional inference forests, Ranger, and survival forests with $L_1$ splitting, with two novel variants, namely distributional and transformation survival forests. Theoretical considerations explain the low power of log-rank-based splitting in detecting patterns in non-proportional hazards situations in survival trees and corresponding forests. This limitation can potentially be overcome by the alternative split procedures suggested herein. We empirically investigated this effect using simulation experiments and a re-analysis of the PRO-ACT database of ALS survival, giving special emphasis to both prognostic and predictive models.
OpenML: An R Package to Connect to the Machine Learning Platform OpenML
May 04, 2017



Abstract:OpenML is an online machine learning platform where researchers can easily share data, machine learning tasks and experiments as well as organize them online to work and collaborate more efficiently. In this paper, we present an R package to interface with the OpenML platform and illustrate its usage in combination with the machine learning R package mlr. We show how the OpenML package allows R users to easily search, download and upload data sets and machine learning tasks. Furthermore, we also show how to upload results of experiments, share them with others and download results from other users. Beyond ensuring reproducibility of results, the OpenML platform automates much of the drudge work, speeds up research, facilitates collaboration and increases the users' visibility online.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge