What Makes Forest-Based Heterogeneous Treatment Effect Estimators Work?
Paper and Code
Jun 21, 2022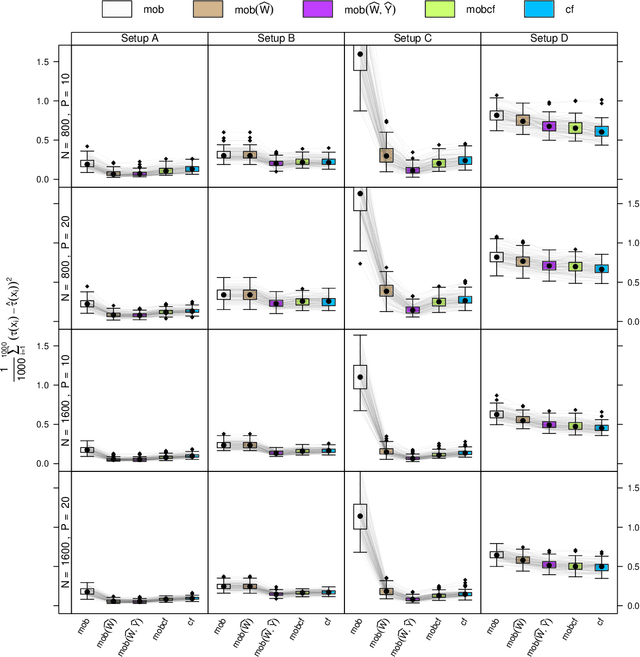
Estimation of heterogeneous treatment effects (HTE) is of prime importance in many disciplines, ranging from personalized medicine to economics among many others. Random forests have been shown to be a flexible and powerful approach to HTE estimation in both randomized trials and observational studies. In particular "causal forests", introduced by Athey, Tibshirani and Wager (2019), along with the R implementation in package grf were rapidly adopted. A related approach, called "model-based forests", that is geared towards randomized trials and simultaneously captures effects of both prognostic and predictive variables, was introduced by Seibold, Zeileis and Hothorn (2018) along with a modular implementation in the R package model4you. Here, we present a unifying view that goes beyond the theoretical motivations and investigates which computational elements make causal forests so successful and how these can be blended with the strengths of model-based forests. To do so, we show that both methods can be understood in terms of the same parameters and model assumptions for an additive model under L2 loss. This theoretical insight allows us to implement several flavors of "model-based causal forests" and dissect their different elements in silico. The original causal forests and model-based forests are compared with the new blended versions in a benchmark study exploring both randomized trials and observational settings. In the randomized setting, both approaches performed akin. If confounding was present in the data generating process, we found local centering of the treatment indicator with the corresponding propensities to be the main driver for good performance. Local centering of the outcome was less important, and might be replaced or enhanced by simultaneous split selection with respect to both prognostic and predictive effects.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge