Hassan Bagher Ebadian
MulModSeg: Enhancing Unpaired Multi-Modal Medical Image Segmentation with Modality-Conditioned Text Embedding and Alternating Training
Nov 23, 2024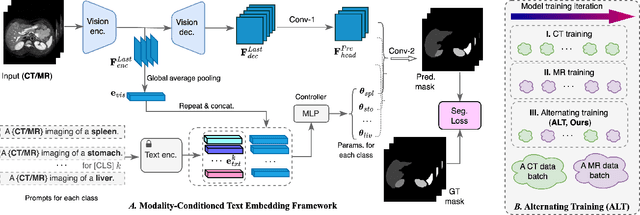
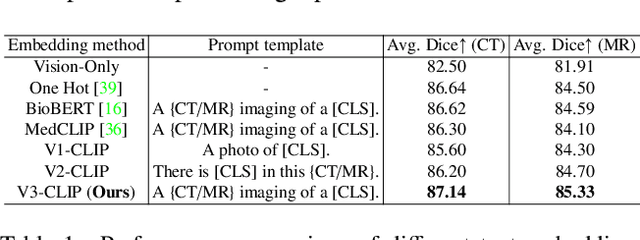
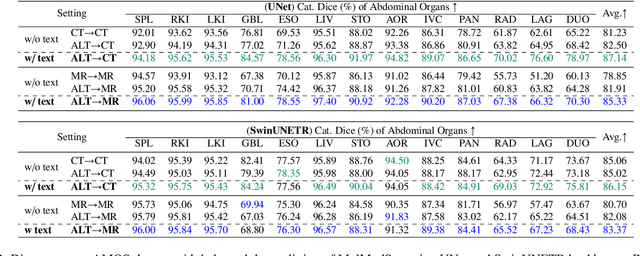
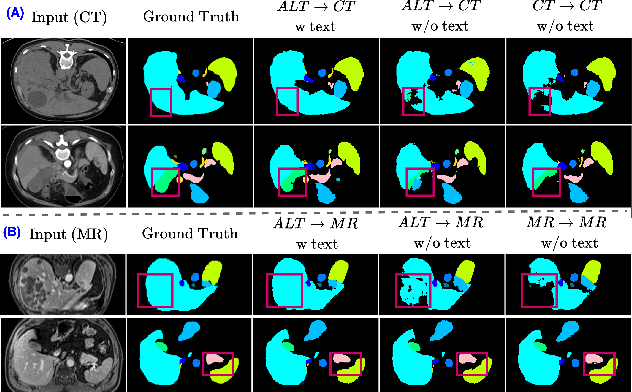
Abstract:In the diverse field of medical imaging, automatic segmentation has numerous applications and must handle a wide variety of input domains, such as different types of Computed Tomography (CT) scans and Magnetic Resonance (MR) images. This heterogeneity challenges automatic segmentation algorithms to maintain consistent performance across different modalities due to the requirement for spatially aligned and paired images. Typically, segmentation models are trained using a single modality, which limits their ability to generalize to other types of input data without employing transfer learning techniques. Additionally, leveraging complementary information from different modalities to enhance segmentation precision often necessitates substantial modifications to popular encoder-decoder designs, such as introducing multiple branched encoding or decoding paths for each modality. In this work, we propose a simple Multi-Modal Segmentation (MulModSeg) strategy to enhance medical image segmentation across multiple modalities, specifically CT and MR. It incorporates two key designs: a modality-conditioned text embedding framework via a frozen text encoder that adds modality awareness to existing segmentation frameworks without significant structural modifications or computational overhead, and an alternating training procedure that facilitates the integration of essential features from unpaired CT and MR inputs. Through extensive experiments with both Fully Convolutional Network and Transformer-based backbones, MulModSeg consistently outperforms previous methods in segmenting abdominal multi-organ and cardiac substructures for both CT and MR modalities. The code is available in this {\href{https://github.com/ChengyinLee/MulModSeg_2024}{link}}.
Prostate Cancer Malignancy Detection and localization from mpMRI using auto-Deep Learning: One Step Closer to Clinical Utilization
Jun 13, 2022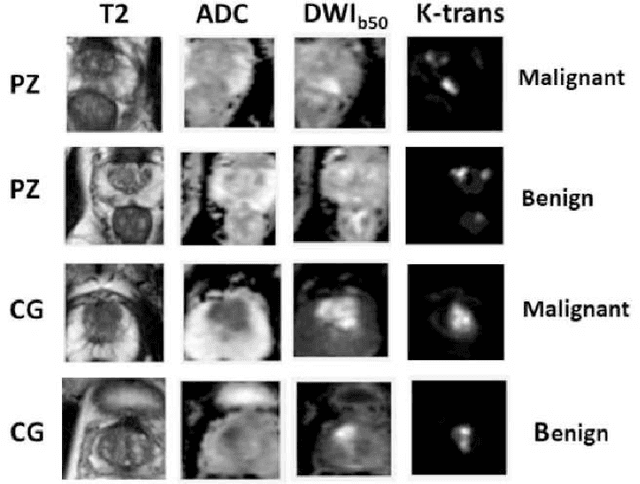

Abstract:Automatic diagnosis of malignant prostate cancer patients from mpMRI has been studied heavily in the past years. Model interpretation and domain drift have been the main road blocks for clinical utilization. As an extension from our previous work where we trained a customized convolutional neural network on a public cohort with 201 patients and the cropped 2D patches around the region of interest were used as the input, the cropped 2.5D slices of the prostate glands were used as the input, and the optimal model were searched in the model space using autoKeras. Something different was peripheral zone (PZ) and central gland (CG) were trained and tested separately, the PZ detector and CG detector were demonstrated effectively in highlighting the most suspicious slices out of a sequence, hopefully to greatly ease the workload for the physicians.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge