Haleh Akrami
Meta Transfer of Self-Supervised Knowledge: Foundation Model in Action for Post-Traumatic Epilepsy Prediction
Dec 21, 2023Abstract:Despite the impressive advancements achieved using deep-learning for functional brain activity analysis, the heterogeneity of functional patterns and scarcity of imaging data still pose challenges in tasks such as prediction of future onset of Post-Traumatic Epilepsy (PTE) from data acquired shortly after traumatic brain injury (TBI). Foundation models pre-trained on separate large-scale datasets can improve the performance from scarce and heterogeneous datasets. For functional Magnetic Resonance Imaging (fMRI), while data may be abundantly available from healthy controls, clinical data is often scarce, limiting the ability of foundation models to identify clinically-relevant features. We overcome this limitation by introducing a novel training strategy for our foundation model by integrating meta-learning with self-supervised learning to improve the generalization from normal to clinical features. In this way we enable generalization to other downstream clinical tasks, in our case prediction of PTE. To achieve this, we perform self-supervised training on the control dataset to focus on inherent features that are not limited to a particular supervised task while applying meta-learning, which strongly improves the model's generalizability using bi-level optimization. Through experiments on neurological disorder classification tasks, we demonstrate that the proposed strategy significantly improves task performance on small-scale clinical datasets. To explore the generalizability of the foundation model in downstream applications, we then apply the model to an unseen TBI dataset for prediction of PTE using zero-shot learning. Results further demonstrated the enhanced generalizability of our foundation model.
Learning A Disentangling Representation For PU Learning
Oct 05, 2023Abstract:In this paper, we address the problem of learning a binary (positive vs. negative) classifier given Positive and Unlabeled data commonly referred to as PU learning. Although rudimentary techniques like clustering, out-of-distribution detection, or positive density estimation can be used to solve the problem in low-dimensional settings, their efficacy progressively deteriorates with higher dimensions due to the increasing complexities in the data distribution. In this paper we propose to learn a neural network-based data representation using a loss function that can be used to project the unlabeled data into two (positive and negative) clusters that can be easily identified using simple clustering techniques, effectively emulating the phenomenon observed in low-dimensional settings. We adopt a vector quantization technique for the learned representations to amplify the separation between the learned unlabeled data clusters. We conduct experiments on simulated PU data that demonstrate the improved performance of our proposed method compared to the current state-of-the-art approaches. We also provide some theoretical justification for our two cluster-based approach and our algorithmic choices.
Beta quantile regression for robust estimation of uncertainty in the presence of outliers
Sep 14, 2023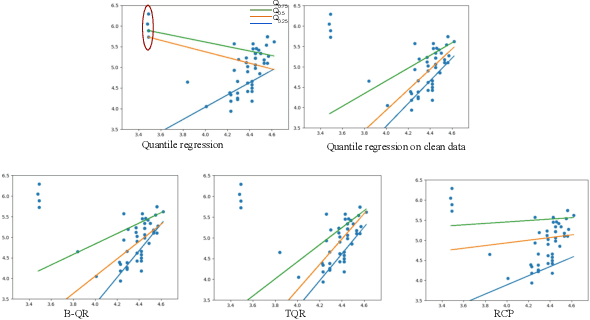
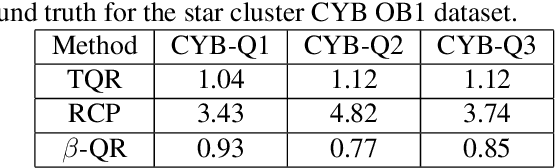
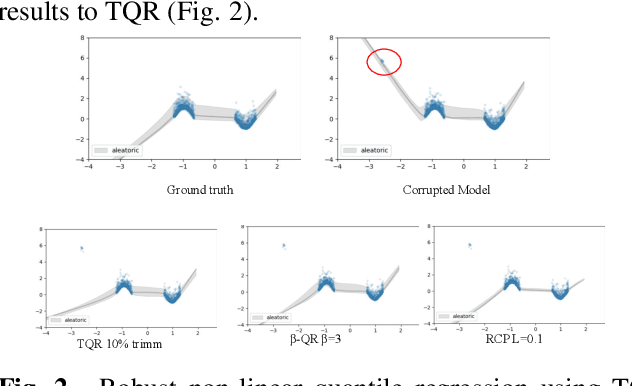
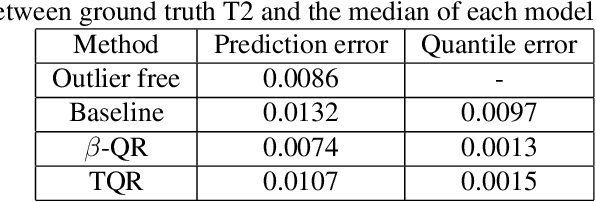
Abstract:Quantile Regression (QR) can be used to estimate aleatoric uncertainty in deep neural networks and can generate prediction intervals. Quantifying uncertainty is particularly important in critical applications such as clinical diagnosis, where a realistic assessment of uncertainty is essential in determining disease status and planning the appropriate treatment. The most common application of quantile regression models is in cases where the parametric likelihood cannot be specified. Although quantile regression is quite robust to outlier response observations, it can be sensitive to outlier covariate observations (features). Outlier features can compromise the performance of deep learning regression problems such as style translation, image reconstruction, and deep anomaly detection, potentially leading to misleading conclusions. To address this problem, we propose a robust solution for quantile regression that incorporates concepts from robust divergence. We compare the performance of our proposed method with (i) least trimmed quantile regression and (ii) robust regression based on the regularization of case-specific parameters in a simple real dataset in the presence of outlier. These methods have not been applied in a deep learning framework. We also demonstrate the applicability of the proposed method by applying it to a medical imaging translation task using diffusion models.
Toward Improved Generalization: Meta Transfer of Self-supervised Knowledge on Graphs
Dec 16, 2022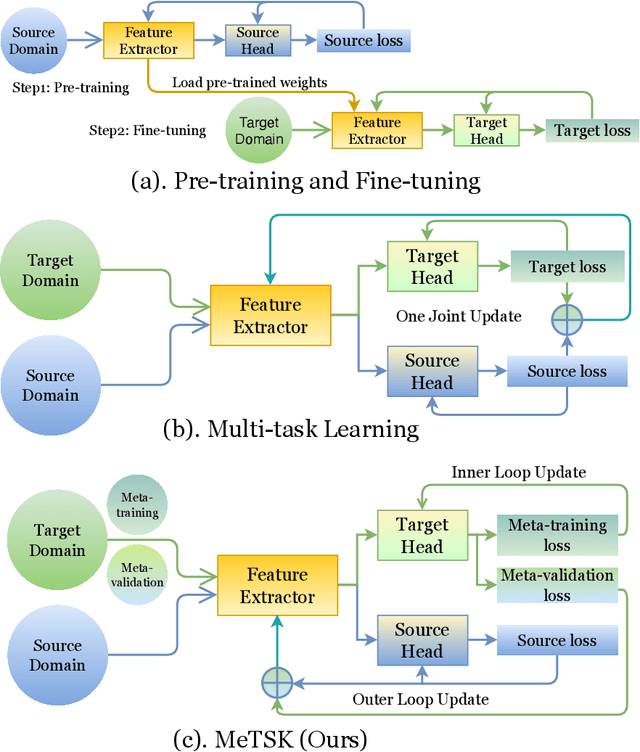

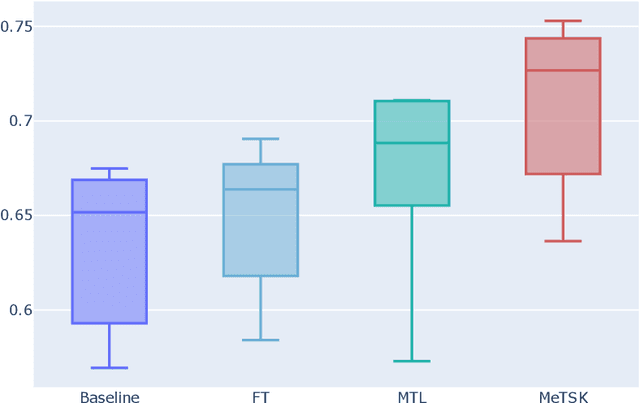

Abstract:Despite the remarkable success achieved by graph convolutional networks for functional brain activity analysis, the heterogeneity of functional patterns and the scarcity of imaging data still pose challenges in many tasks. Transferring knowledge from a source domain with abundant training data to a target domain is effective for improving representation learning on scarce training data. However, traditional transfer learning methods often fail to generalize the pre-trained knowledge to the target task due to domain discrepancy. Self-supervised learning on graphs can increase the generalizability of graph features since self-supervision concentrates on inherent graph properties that are not limited to a particular supervised task. We propose a novel knowledge transfer strategy by integrating meta-learning with self-supervised learning to deal with the heterogeneity and scarcity of fMRI data. Specifically, we perform a self-supervised task on the source domain and apply meta-learning, which strongly improves the generalizability of the model using the bi-level optimization, to transfer the self-supervised knowledge to the target domain. Through experiments on a neurological disorder classification task, we demonstrate that the proposed strategy significantly improves target task performance by increasing the generalizability and transferability of graph-based knowledge.
Speech MOS multi-task learning and rater bias correction
Dec 04, 2022



Abstract:Perceptual speech quality is an important performance metric for teleconferencing applications. The mean opinion score (MOS) is standardized for the perceptual evaluation of speech quality and is obtained by asking listeners to rate the quality of a speech sample. Recently, there has been increasing research interest in developing models for estimating MOS blindly. Here we propose a multi-task framework to include additional labels and data in training to improve the performance of a blind MOS estimation model. Experimental results indicate that the proposed model can be trained to jointly estimate MOS, reverberation time (T60), and clarity (C50) by combining two disjoint data sets in training, one containing only MOS labels and the other containing only T60 and C50 labels. Furthermore, we use a semi-supervised framework to combine two MOS data sets in training, one containing only MOS labels (per ITU-T Recommendation P.808), and the other containing separate scores for speech signal, background noise, and overall quality (per ITU-T Recommendation P.835). Finally, we present preliminary results for addressing individual rater bias in the MOS labels.
Learning From Positive and Unlabeled Data Using Observer-GAN
Aug 26, 2022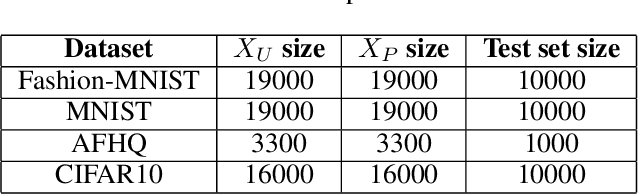



Abstract:The problem of learning from positive and unlabeled data (A.K.A. PU learning) has been studied in a binary (i.e., positive versus negative) classification setting, where the input data consist of (1) observations from the positive class and their corresponding labels, (2) unlabeled observations from both positive and negative classes. Generative Adversarial Networks (GANs) have been used to reduce the problem to the supervised setting with the advantage that supervised learning has state-of-the-art accuracy in classification tasks. In order to generate \textit{pseudo}-negative observations, GANs are trained on positive and unlabeled observations with a modified loss. Using both positive and \textit{pseudo}-negative observations leads to a supervised learning setting. The generation of pseudo-negative observations that are realistic enough to replace missing negative class samples is a bottleneck for current GAN-based algorithms. By including an additional classifier into the GAN architecture, we provide a novel GAN-based approach. In our suggested method, the GAN discriminator instructs the generator only to produce samples that fall into the unlabeled data distribution, while a second classifier (observer) network monitors the GAN training to: (i) prevent the generated samples from falling into the positive distribution; and (ii) learn the features that are the key distinction between the positive and negative observations. Experiments on four image datasets demonstrate that our trained observer network performs better than existing techniques in discriminating between real unseen positive and negative samples.
Learning from imperfect training data using a robust loss function: application to brain image segmentation
Aug 08, 2022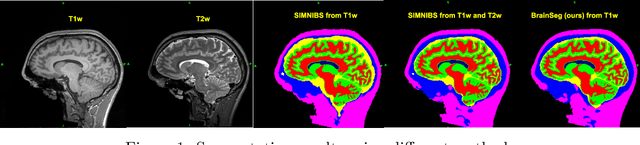
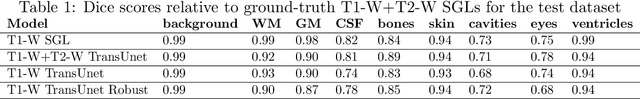
Abstract:Segmentation is one of the most important tasks in MRI medical image analysis and is often the first and the most critical step in many clinical applications. In brain MRI analysis, head segmentation is commonly used for measuring and visualizing the brain's anatomical structures and is also a necessary step for other applications such as current-source reconstruction in electroencephalography and magnetoencephalography (EEG/MEG). Here we propose a deep learning framework that can segment brain, skull, and extra-cranial tissue using only T1-weighted MRI as input. In addition, we describe a robust method for training the model in the presence of noisy labels.
Semi-supervised Learning using Robust Loss
Mar 03, 2022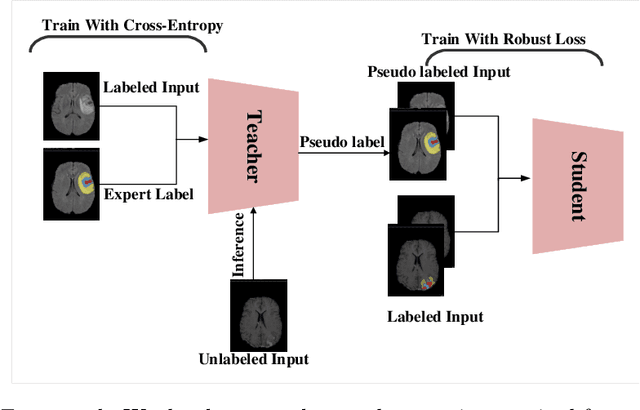
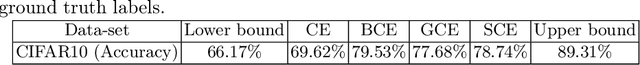
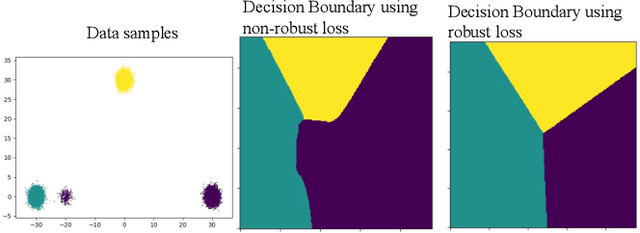
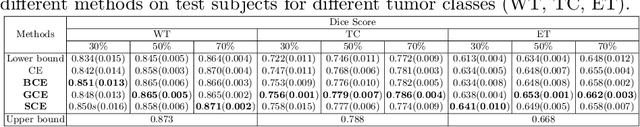
Abstract:The amount of manually labeled data is limited in medical applications, so semi-supervised learning and automatic labeling strategies can be an asset for training deep neural networks. However, the quality of the automatically generated labels can be uneven and inferior to manual labels. In this paper, we suggest a semi-supervised training strategy for leveraging both manually labeled data and extra unlabeled data. In contrast to the existing approaches, we apply robust loss for the automated labeled data to automatically compensate for the uneven data quality using a teacher-student framework. First, we generate pseudo-labels for unlabeled data using a teacher model pre-trained on labeled data. These pseudo-labels are noisy, and using them along with labeled data for training a deep neural network can severely degrade learned feature representations and the generalization of the network. Here we mitigate the effect of these pseudo-labels by using robust loss functions. Specifically, we use three robust loss functions, namely beta cross-entropy, symmetric cross-entropy, and generalized cross-entropy. We show that our proposed strategy improves the model performance by compensating for the uneven quality of labels in image classification as well as segmentation applications.
Deep Quantile Regression for Uncertainty Estimation in Unsupervised and Supervised Lesion Detection
Sep 20, 2021



Abstract:Despite impressive state-of-the-art performance on a wide variety of machine learning tasks in multiple applications, deep learning methods can produce over-confident predictions, particularly with limited training data. Therefore, quantifying uncertainty is particularly important in critical applications such as anomaly or lesion detection and clinical diagnosis, where a realistic assessment of uncertainty is essential in determining surgical margins, disease status and appropriate treatment. In this work, we focus on using quantile regression to estimate aleatoric uncertainty and use it for estimating uncertainty in both supervised and unsupervised lesion detection problems. In the unsupervised settings, we apply quantile regression to a lesion detection task using Variational AutoEncoder (VAE). The VAE models the output as a conditionally independent Gaussian characterized by means and variances for each output dimension. Unfortunately, joint optimization of both mean and variance in the VAE leads to the well-known problem of shrinkage or underestimation of variance. We describe an alternative VAE model, Quantile-Regression VAE (QR-VAE), that avoids this variance shrinkage problem by estimating conditional quantiles for the given input image. Using the estimated quantiles, we compute the conditional mean and variance for input images under the conditionally Gaussian model. We then compute reconstruction probability using this model as a principled approach to outlier or anomaly detection applications. In the supervised setting, we develop binary quantile regression (BQR) for the supervised lesion segmentation task. BQR segmentation can capture uncertainty in label boundaries. We show how quantile regression can be used to characterize expert disagreement in the location of lesion boundaries.
fMRI-Kernel Regression: A Kernel-based Method for Pointwise Statistical Analysis of rs-fMRI for Population Studies
Dec 13, 2020



Abstract:Due to the spontaneous nature of resting-state fMRI (rs-fMRI) signals, cross-subject comparison and therefore, group studies of rs-fMRI are challenging. Most existing group comparison methods use features extracted from the fMRI time series, such as connectivity features, independent component analysis (ICA), and functional connectivity density (FCD) methods. However, in group studies, especially in the case of spectrum disorders, distances to a single atlas or a representative subject do not fully reflect the differences between subjects that may lie on a multi-dimensional spectrum. Moreover, there may not exist an individual subject or even an average atlas in such cases that is representative of all subjects. Here we describe an approach that measures pairwise distances between the synchronized rs-fMRI signals of pairs of subjects instead of to a single reference point. We also present a method for fMRI data comparison that leverages this generated pairwise feature to establish a radial basis function kernel matrix. This kernel matrix is used in turn to perform kernel regression of rs-fMRI to a clinical variable such as a cognitive or neurophysiological performance score of interest. This method opens a new pointwise analysis paradigm for fMRI data. We demonstrate the application of this method by performing a pointwise analysis on the cortical surface using rs-fMRI data to identify cortical regions associated with variability in ADHD index. While pointwise analysis methods are common in anatomical studies such as cortical thickness analysis and voxel- and tensor-based morphometry and its variants, such a method is lacking for rs-fMRI and could improve the utility of rs-fMRI for group studies. The method presented in this paper is aimed at filling this gap.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge