Guozheng Xu
RoofSeg: An edge-aware transformer-based network for end-to-end roof plane segmentation
Aug 26, 2025Abstract:Roof plane segmentation is one of the key procedures for reconstructing three-dimensional (3D) building models at levels of detail (LoD) 2 and 3 from airborne light detection and ranging (LiDAR) point clouds. The majority of current approaches for roof plane segmentation rely on the manually designed or learned features followed by some specifically designed geometric clustering strategies. Because the learned features are more powerful than the manually designed features, the deep learning-based approaches usually perform better than the traditional approaches. However, the current deep learning-based approaches have three unsolved problems. The first is that most of them are not truly end-to-end, the plane segmentation results may be not optimal. The second is that the point feature discriminability near the edges is relatively low, leading to inaccurate planar edges. The third is that the planar geometric characteristics are not sufficiently considered to constrain the network training. To solve these issues, a novel edge-aware transformer-based network, named RoofSeg, is developed for segmenting roof planes from LiDAR point clouds in a truly end-to-end manner. In the RoofSeg, we leverage a transformer encoder-decoder-based framework to hierarchically predict the plane instance masks with the use of a set of learnable plane queries. To further improve the segmentation accuracy of edge regions, we also design an Edge-Aware Mask Module (EAMM) that sufficiently incorporates planar geometric prior of edges to enhance its discriminability for plane instance mask refinement. In addition, we propose an adaptive weighting strategy in the mask loss to reduce the influence of misclassified points, and also propose a new plane geometric loss to constrain the network training.
A boundary-aware point clustering approach in Euclidean and embedding spaces for roof plane segmentation
Sep 07, 2023Abstract:Roof plane segmentation from airborne LiDAR point clouds is an important technology for 3D building model reconstruction. One of the key issues of plane segmentation is how to design powerful features that can exactly distinguish adjacent planar patches. The quality of point feature directly determines the accuracy of roof plane segmentation. Most of existing approaches use handcrafted features to extract roof planes. However, the abilities of these features are relatively low, especially in boundary area. To solve this problem, we propose a boundary-aware point clustering approach in Euclidean and embedding spaces constructed by a multi-task deep network for roof plane segmentation. We design a three-branch network to predict semantic labels, point offsets and extract deep embedding features. In the first branch, we classify the input data as non-roof, boundary and plane points. In the second branch, we predict point offsets for shifting each point toward its respective instance center. In the third branch, we constrain that points of the same plane instance should have the similar embeddings. We aim to ensure that points of the same plane instance are close as much as possible in both Euclidean and embedding spaces. However, although deep network has strong feature representative ability, it is still hard to accurately distinguish points near plane instance boundary. Therefore, we first group plane points into many clusters in the two spaces, and then we assign the rest boundary points to their closest clusters to generate final complete roof planes. In this way, we can effectively reduce the influence of unreliable boundary points. In addition, we construct a synthetic dataset and a real dataset to train and evaluate our approach. The experiments results show that the proposed approach significantly outperforms the existing state-of-the-art approaches.
Reinventing 2D Convolutions for 3D Medical Images
Nov 24, 2019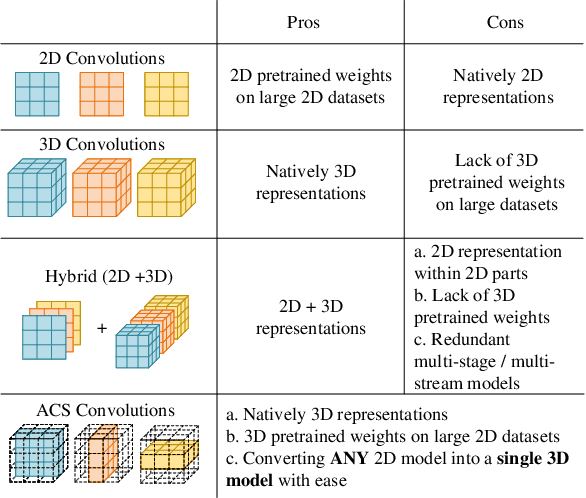
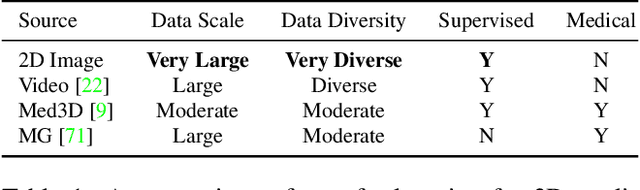
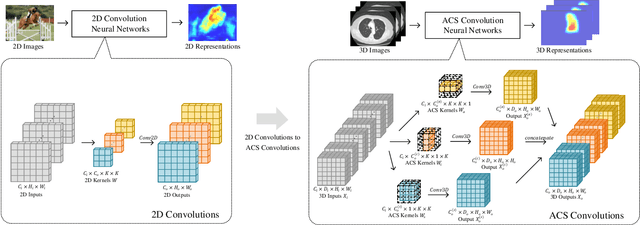
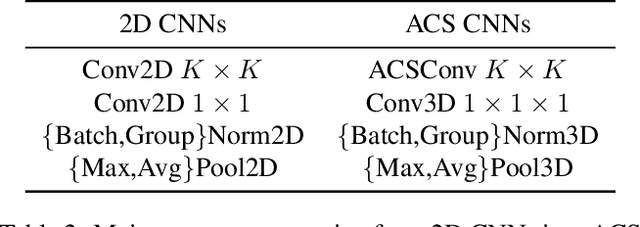
Abstract:There has been considerable debate over 2D and 3D representation learning on 3D medical images. 2D approaches could benefit from large-scale 2D pretraining, whereas they are generally weak in capturing large 3D contexts. 3D approaches are natively strong in 3D contexts, however few publicly available 3D medical dataset is large and diverse enough for universal 3D pretraining. Even for hybrid (2D + 3D) approaches, the intrinsic disadvantages within the 2D / 3D parts still exist. In this study, we bridge the gap between 2D and 3D convolutions by reinventing the 2D convolutions. We propose ACS (axial-coronal-sagittal) convolutions to perform natively 3D representation learning, while utilizing the pretrained weights from 2D counterparts. In ACS convolutions, 2D convolution kernels are split by channel into three parts, and convoluted separately on the three views (axial, coronal and sagittal) of 3D representations. Theoretically, ANY 2D CNN (ResNet, DenseNet, or DeepLab) is able to be converted into a 3D ACS CNN, with pretrained weights of same parameter sizes. Extensive experiments on proof-of-concept dataset and several medical benchmarks validate the consistent superiority of the pretrained ACS CNNs, over the 2D / 3D CNN counterparts with / without pretraining. Even without pretraining, the ACS convolution can be used as a plug-and-play replacement of standard 3D convolution, with smaller model size.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge