Guliz Irem Gokceler
Deep Learning-based Frozen Section to FFPE Translation
Jul 27, 2021



Abstract:Frozen sectioning (FS) is the preparation method of choice for microscopic evaluation of tissues during surgical operations. The high speed of the procedure allows pathologists to rapidly assess the key microscopic features, such as tumour margins and malignant status to guide surgical decision-making and minimise disruptions to the course of the operation. However, FS is prone to introducing many misleading artificial structures (histological artefacts), such as nuclear ice crystals, compression, and cutting artefacts, hindering timely and accurate diagnostic judgement of the pathologist. Additional training and prolonged experience is often required to make highly effective and time-critical diagnosis on frozen sections. On the other hand, the gold standard tissue preparation technique of formalin-fixation and paraffin-embedding (FFPE) provides significantly superior image quality, but is a very time-consuming process (12-48 hours), making it unsuitable for intra-operative use. In this paper, we propose an artificial intelligence (AI) method that improves FS image quality by computationally transforming frozen-sectioned whole-slide images (FS-WSIs) into whole-slide FFPE-style images in minutes. AI-FFPE rectifies FS artefacts with the guidance of an attention mechanism that puts a particular emphasis on artefacts while utilising a self-regularization mechanism established between FS input image and synthesized FFPE-style image that preserves clinically relevant features. As a result, AI-FFPE method successfully generates FFPE-style images without significantly extending tissue processing time and consequently improves diagnostic accuracy. We demonstrate the efficacy of AI-FFPE on lung and brain frozen sections using a variety of different qualitative and quantitative metrics including visual Turing tests from 20 board certified pathologists.
VR-Caps: A Virtual Environment for Capsule Endoscopy
Aug 29, 2020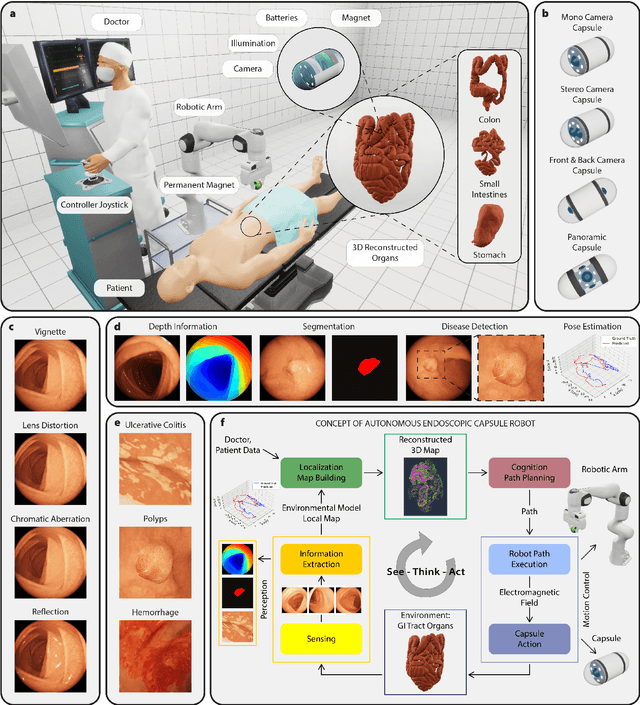
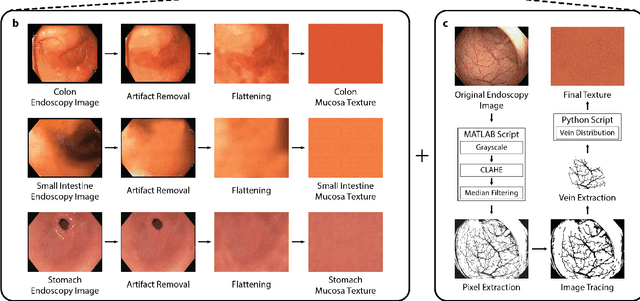
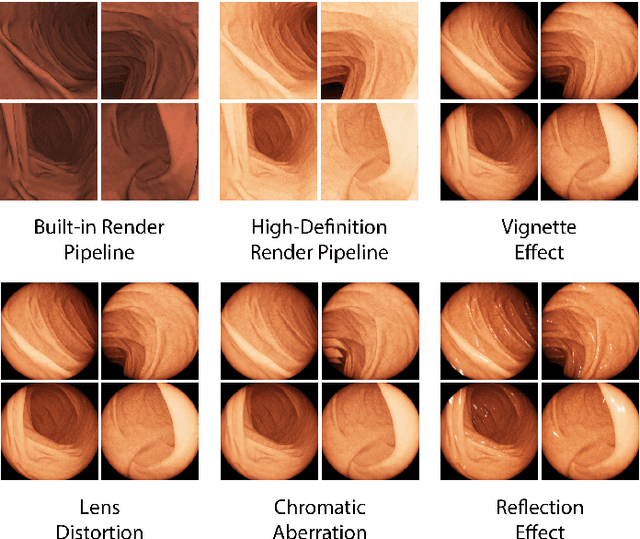
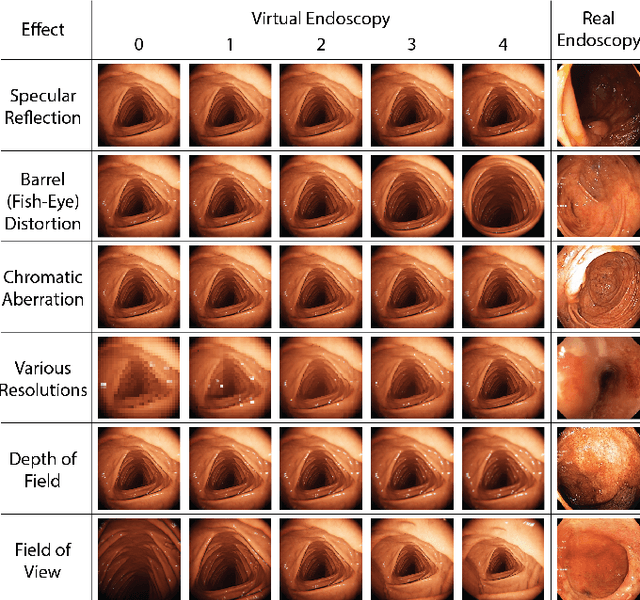
Abstract:Current capsule endoscopes and next-generation robotic capsules for diagnosis and treatment of gastrointestinal diseases are complex cyber-physical platforms that must orchestrate complex software and hardware functions. The desired tasks for these systems include visual localization, depth estimation, 3D mapping, disease detection and segmentation, automated navigation, active control, path realization and optional therapeutic modules such as targeted drug delivery and biopsy sampling. Data-driven algorithms promise to enable many advanced functionalities for capsule endoscopes, but real-world data is challenging to obtain. Physically-realistic simulations providing synthetic data have emerged as a solution to the development of data-driven algorithms. In this work, we present a comprehensive simulation platform for capsule endoscopy operations and introduce VR-Caps, a virtual active capsule environment that simulates a range of normal and abnormal tissue conditions (e.g., inflated, dry, wet etc.) and varied organ types, capsule endoscope designs (e.g., mono, stereo, dual and 360{\deg}camera), and the type, number, strength, and placement of internal and external magnetic sources that enable active locomotion. VR-Caps makes it possible to both independently or jointly develop, optimize, and test medical imaging and analysis software for the current and next-generation endoscopic capsule systems. To validate this approach, we train state-of-the-art deep neural networks to accomplish various medical image analysis tasks using simulated data from VR-Caps and evaluate the performance of these models on real medical data. Results demonstrate the usefulness and effectiveness of the proposed virtual platform in developing algorithms that quantify fractional coverage, camera trajectory, 3D map reconstruction, and disease classification.
Quantitative Evaluation of Endoscopic SLAM Methods: EndoSLAM Dataset
Jul 01, 2020

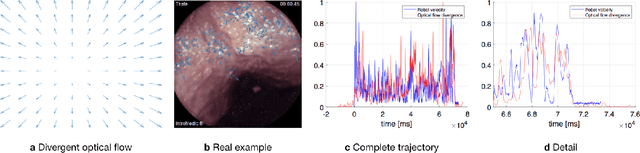
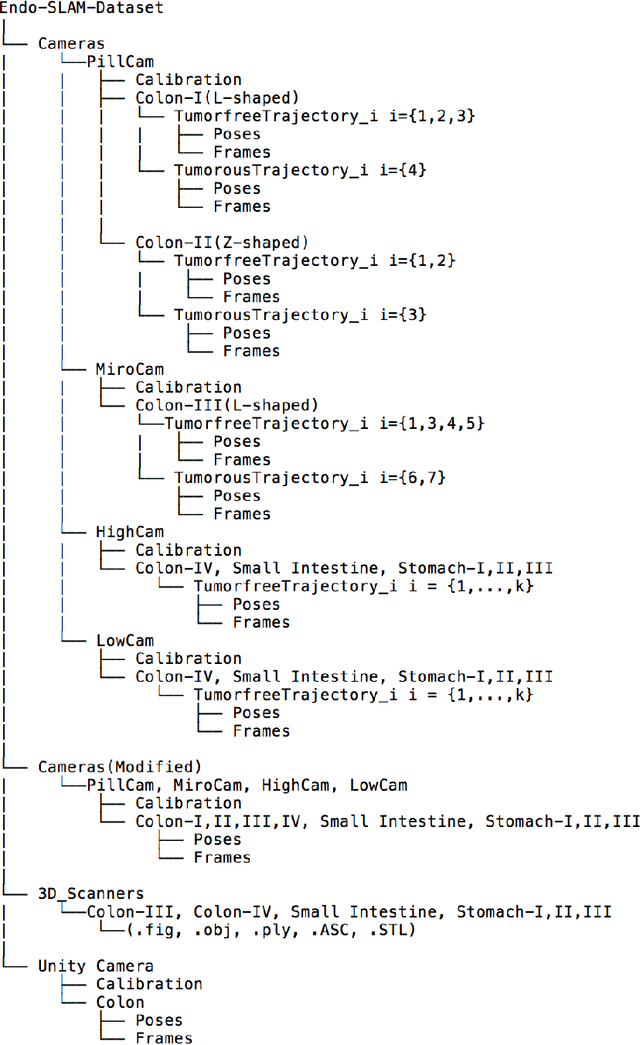
Abstract:Deep learning techniques hold promise to improve dense topography reconstruction and pose estimation, as well as simultaneous localization and mapping (SLAM). However, currently available datasets do not support effective quantitative benchmarking. With this paper, we introduce a comprehensive endoscopic SLAM dataset containing both capsule and standard endoscopy recordings. A Panda robotic arm, two different commercially available high precision 3D scanners, two different commercially available capsule endoscopes with different camera properties and two different conventional endoscopy cameras were employed to collect data from eight ex-vivo porcine gastrointestinal (GI)-tract organs. In total, 35 sub-datasets are provided: 18 sub-datasets for colon, 12 sub-datasets for stomach and five sub-datasets for small intestine, while four of these contain polyp-mimicking elevations carried out by an expert gastroenterologist. To exemplify the use-case, SC-SfMLearner was comprehensively benchmarked. The codes and the link for the dataset are publicly available at https://github.com/CapsuleEndoscope/EndoSLAM. A video demonstrating the experimental setup and procedure is available at https://www.youtube.com/watch?v=G_LCe0aWWdQ.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge