Guanjun Wang
PHAT: Modeling Period Heterogeneity for Multivariate Time Series Forecasting
Jan 31, 2026Abstract:While existing multivariate time series forecasting models have advanced significantly in modeling periodicity, they largely neglect the periodic heterogeneity common in real-world data, where variates exhibit distinct and dynamically changing periods. To effectively capture this periodic heterogeneity, we propose PHAT (Period Heterogeneity-Aware Transformer). Specifically, PHAT arranges multivariate inputs into a three-dimensional "periodic bucket" tensor, where the dimensions correspond to variate group characteristics with similar periodicity, time steps aligned by phase, and offsets within the period. By restricting interactions within buckets and masking cross-bucket connections, PHAT effectively avoids interference from inconsistent periods. We also propose a positive-negative attention mechanism, which captures periodic dependencies from two perspectives: periodic alignment and periodic deviation. Additionally, the periodic alignment attention scores are decomposed into positive and negative components, with a modulation term encoding periodic priors. This modulation constrains the attention mechanism to more faithfully reflect the underlying periodic trends. A mathematical explanation is provided to support this property. We evaluate PHAT comprehensively on 14 real-world datasets against 18 baselines, and the results show that it significantly outperforms existing methods, achieving highly competitive forecasting performance. Our sources is available at GitHub.
Spatiotemporal Causal Decoupling Model for Air Quality Forecasting
May 26, 2025Abstract:Due to the profound impact of air pollution on human health, livelihoods, and economic development, air quality forecasting is of paramount significance. Initially, we employ the causal graph method to scrutinize the constraints of existing research in comprehensively modeling the causal relationships between the air quality index (AQI) and meteorological features. In order to enhance prediction accuracy, we introduce a novel air quality forecasting model, AirCade, which incorporates a causal decoupling approach. AirCade leverages a spatiotemporal module in conjunction with knowledge embedding techniques to capture the internal dynamics of AQI. Subsequently, a causal decoupling module is proposed to disentangle synchronous causality from past AQI and meteorological features, followed by the dissemination of acquired knowledge to future time steps to enhance performance. Additionally, we introduce a causal intervention mechanism to explicitly represent the uncertainty of future meteorological features, thereby bolstering the model's robustness. Our evaluation of AirCade on an open-source air quality dataset demonstrates over 20\% relative improvement over state-of-the-art models.
Boosting Sclera Segmentation through Semi-supervised Learning with Fewer Labels
Jan 13, 2025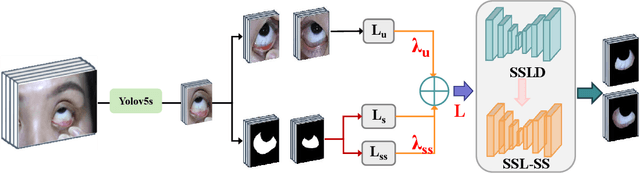

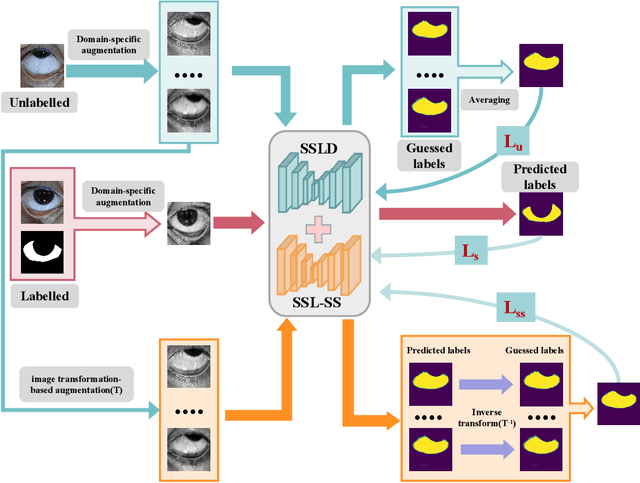
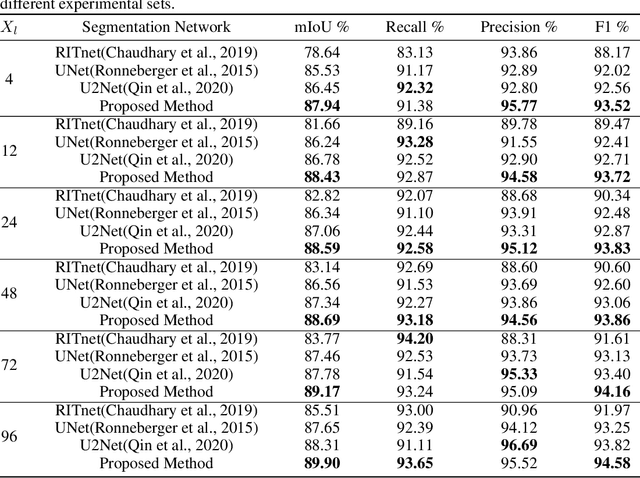
Abstract:Sclera segmentation is crucial for developing automatic eye-related medical computer-aided diagnostic systems, as well as for personal identification and verification, because the sclera contains distinct personal features. Deep learning-based sclera segmentation has achieved significant success compared to traditional methods that rely on hand-crafted features, primarily because it can autonomously extract critical output-related features without the need to consider potential physical constraints. However, achieving accurate sclera segmentation using these methods is challenging due to the scarcity of high-quality, fully labeled datasets, which depend on costly, labor-intensive medical acquisition and expertise. To address this challenge, this paper introduces a novel sclera segmentation framework that excels with limited labeled samples. Specifically, we employ a semi-supervised learning method that integrates domain-specific improvements and image-based spatial transformations to enhance segmentation performance. Additionally, we have developed a real-world eye diagnosis dataset to enrich the evaluation process. Extensive experiments on our dataset and two additional public datasets demonstrate the effectiveness and superiority of our proposed method, especially with significantly fewer labeled samples.
Federated Distillation for Medical Image Classification: Towards Trustworthy Computer-Aided Diagnosis
Jul 03, 2024



Abstract:Medical image classification plays a crucial role in computer-aided clinical diagnosis. While deep learning techniques have significantly enhanced efficiency and reduced costs, the privacy-sensitive nature of medical imaging data complicates centralized storage and model training. Furthermore, low-resource healthcare organizations face challenges related to communication overhead and efficiency due to increasing data and model scales. This paper proposes a novel privacy-preserving medical image classification framework based on federated learning to address these issues, named FedMIC. The framework enables healthcare organizations to learn from both global and local knowledge, enhancing local representation of private data despite statistical heterogeneity. It provides customized models for organizations with diverse data distributions while minimizing communication overhead and improving efficiency without compromising performance. Our FedMIC enhances robustness and practical applicability under resource-constrained conditions. We demonstrate FedMIC's effectiveness using four public medical image datasets for classical medical image classification tasks.
CMT: Interpretable Model for Rapid Recognition Pneumonia from Chest X-Ray Images by Fusing Low Complexity Multilevel Attention Mechanism
Oct 29, 2022



Abstract:Chest imaging plays an essential role in diagnosing and predicting patients with COVID-19 with evidence of worsening respiratory status. Many deep learning-based diagnostic models for pneumonia have been developed to enable computer-aided diagnosis. However, the long training and inference time make them inflexible. In addition, the lack of interpretability reduces their credibility in clinical medical practice. This paper presents CMT, a model with interpretability and rapid recognition of pneumonia, especially COVID-19 positive. Multiple convolutional layers in CMT are first used to extract features in CXR images, and then Transformer is applied to calculate the possibility of each symptom. To improve the model's generalization performance and to address the problem of sparse medical image data, we propose Feature Fusion Augmentation (FFA), a plug-and-play method for image augmentation. It fuses the features of the two images to varying degrees to produce a new image that does not deviate from the original distribution. Furthermore, to reduce the computational complexity and accelerate the convergence, we propose Multilevel Multi-Head Self-Attention (MMSA), which computes attention on different levels to establish the relationship between global and local features. It significantly improves the model performance while substantially reducing its training and inference time. Experimental results on the largest COVID-19 dataset show the proposed CMT has state-of-the-art performance. The effectiveness of FFA and MMSA is demonstrated in the ablation experiments. In addition, the weights and feature activation maps of the model inference process are visualized to show the CMT's interpretability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge