Ghislain Durif
Kernel-Based Testing for Single-Cell Differential Analysis
Jul 17, 2023Abstract:Single-cell technologies have provided valuable insights into the distribution of molecular features, such as gene expression and epigenomic modifications. However, comparing these complex distributions in a controlled and powerful manner poses methodological challenges. Here we propose to benefit from the kernel-testing framework to compare the complex cell-wise distributions of molecular features in a non-linear manner based on their kernel embedding. Our framework not only allows for feature-wise analyses but also enables global comparisons of transcriptomes or epigenomes, considering their intricate dependencies. By using a classifier to discriminate cells based on the variability of their embedding, our method uncovers heterogeneities in cell populations that would otherwise go undetected. We show that kernel testing overcomes the limitations of differential analysis methods dedicated to single-cell. Kernel testing is applied to investigate the reversion process of differentiating cells, successfully identifying cells in transition between reversion and differentiation stages. Additionally, we analyze single-cell ChIP-Seq data and identify a subpopulation of untreated breast cancer cells that exhibit an epigenomic profile similar to persister cells.
Benchopt: Reproducible, efficient and collaborative optimization benchmarks
Jun 28, 2022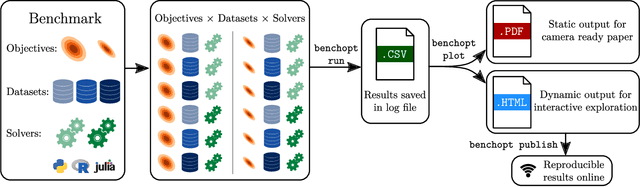
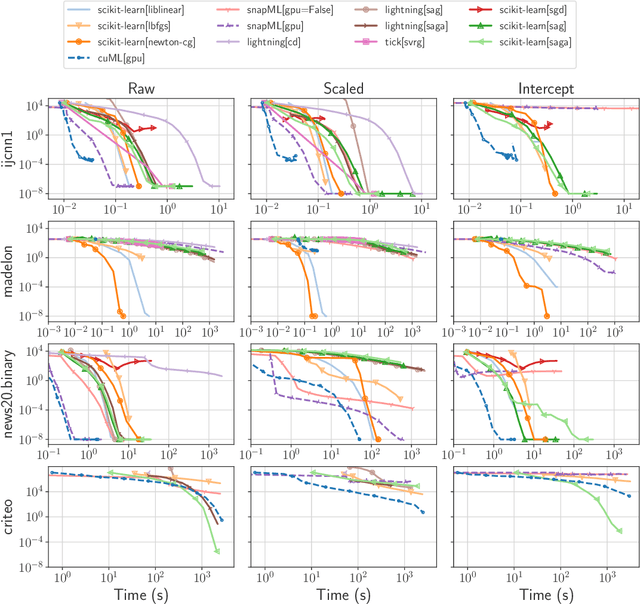
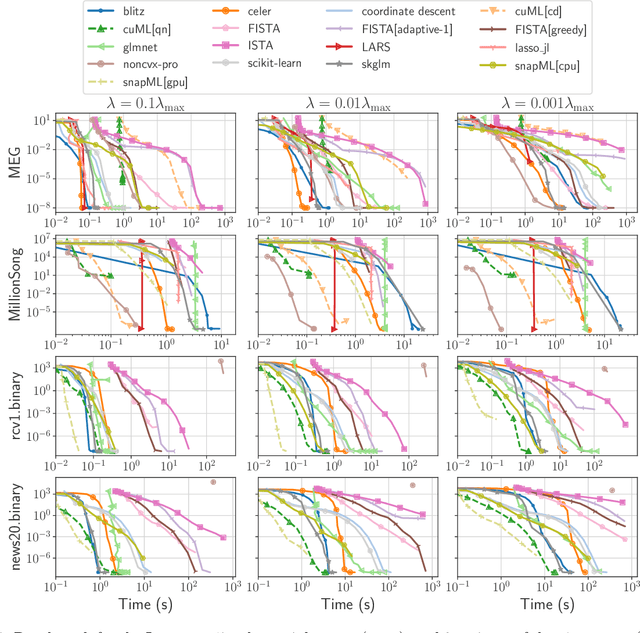
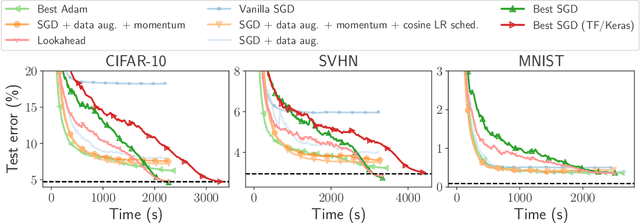
Abstract:Numerical validation is at the core of machine learning research as it allows to assess the actual impact of new methods, and to confirm the agreement between theory and practice. Yet, the rapid development of the field poses several challenges: researchers are confronted with a profusion of methods to compare, limited transparency and consensus on best practices, as well as tedious re-implementation work. As a result, validation is often very partial, which can lead to wrong conclusions that slow down the progress of research. We propose Benchopt, a collaborative framework to automate, reproduce and publish optimization benchmarks in machine learning across programming languages and hardware architectures. Benchopt simplifies benchmarking for the community by providing an off-the-shelf tool for running, sharing and extending experiments. To demonstrate its broad usability, we showcase benchmarks on three standard learning tasks: $\ell_2$-regularized logistic regression, Lasso, and ResNet18 training for image classification. These benchmarks highlight key practical findings that give a more nuanced view of the state-of-the-art for these problems, showing that for practical evaluation, the devil is in the details. We hope that Benchopt will foster collaborative work in the community hence improving the reproducibility of research findings.
Kernel Operations on the GPU, with Autodiff, without Memory Overflows
Mar 27, 2020Abstract:The KeOps library provides a fast and memory-efficient GPU support for tensors whose entries are given by a mathematical formula, such as kernel and distance matrices. KeOps alleviates the major bottleneck of tensor-centric libraries for kernel and geometric applications: memory consumption. It also supports automatic differentiation and outperforms standard GPU baselines, including PyTorch CUDA tensors or the Halide and TVM libraries. KeOps combines optimized C++/CUDA schemes with binders for high-level languages: Python (Numpy and PyTorch), Matlab and GNU R. As a result, high-level "quadratic" codes can now scale up to large data sets with millions of samples processed in seconds. KeOps brings graphics-like performances for kernel methods and is freely available on standard repositories (PyPi, CRAN). To showcase its versatility, we provide tutorials in a wide range of settings online at \url{www.kernel-operations.io}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge