Gen Shi
COMMA: Coordinate-aware Modulated Mamba Network for 3D Dispersed Vessel Segmentation
Mar 04, 2025



Abstract:Accurate segmentation of 3D vascular structures is essential for various medical imaging applications. The dispersed nature of vascular structures leads to inherent spatial uncertainty and necessitates location awareness, yet most current 3D medical segmentation models rely on the patch-wise training strategy that usually loses this spatial context. In this study, we introduce the Coordinate-aware Modulated Mamba Network (COMMA) and contribute a manually labeled dataset of 570 cases, the largest publicly available 3D vessel dataset to date. COMMA leverages both entire and cropped patch data through global and local branches, ensuring robust and efficient spatial location awareness. Specifically, COMMA employs a channel-compressed Mamba (ccMamba) block to encode entire image data, capturing long-range dependencies while optimizing computational costs. Additionally, we propose a coordinate-aware modulated (CaM) block to enhance interactions between the global and local branches, allowing the local branch to better perceive spatial information. We evaluate COMMA on six datasets, covering two imaging modalities and five types of vascular tissues. The results demonstrate COMMA's superior performance compared to state-of-the-art methods with computational efficiency, especially in segmenting small vessels. Ablation studies further highlight the importance of our proposed modules and spatial information. The code and data will be open source at https://github.com/shigen-StoneRoot/COMMA.
FSC-loss: A Frequency-domain Structure Consistency Learning Approach for Signal Data Recovery and Reconstruction
Jan 08, 2025



Abstract:A core challenge for signal data recovery is to model the distribution of signal matrix (SM) data based on measured low-quality data in biomedical engineering of magnetic particle imaging (MPI). For acquiring the high-resolution (high-quality) SM, the number of meticulous measurements at numerous positions in the field-of-view proves time-consuming (measurement of a 37x37x37 SM takes about 32 hours). To improve reconstructed signal quality and shorten SM measurement time, existing methods explore to generating high-resolution SM based on time-saving measured low-resolution SM (a 9x9x9 SM just takes about 0.5 hours). However, previous methods show poor performance for high-frequency signal recovery in SM. To achieve a high-resolution SM recovery and shorten its acquisition time, we propose a frequency-domain structure consistency loss function and data component embedding strategy to model global and local structural information of SM. We adopt a transformer-based network to evaluate this function and the strategy. We evaluate our methods and state-of-the-art (SOTA) methods on the two simulation datasets and four public measured SMs in Open MPI Data. The results show that our method outperforms the SOTA methods in high-frequency structural signal recovery. Additionally, our method can recover a high-resolution SM with clear high-frequency structure based on a down-sampling factor of 16 less than 15 seconds, which accelerates the acquisition time over 60 times faster than the measurement-based HR SM with the minimum error (nRMSE=0.041). Moreover, our method is applied in our three in-house MPI systems, and boost their performance for signal reconstruction.
Benefit from public unlabeled data: A Frangi filtering-based pretraining network for 3D cerebrovascular segmentation
Dec 23, 2023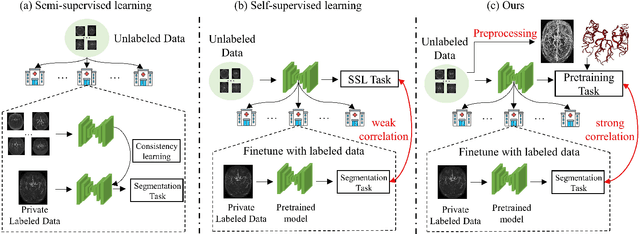
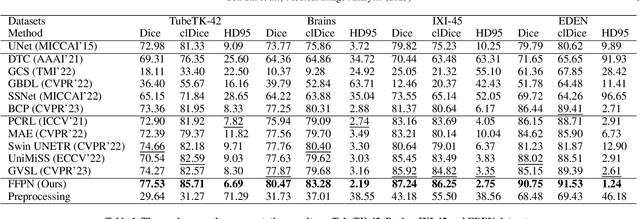
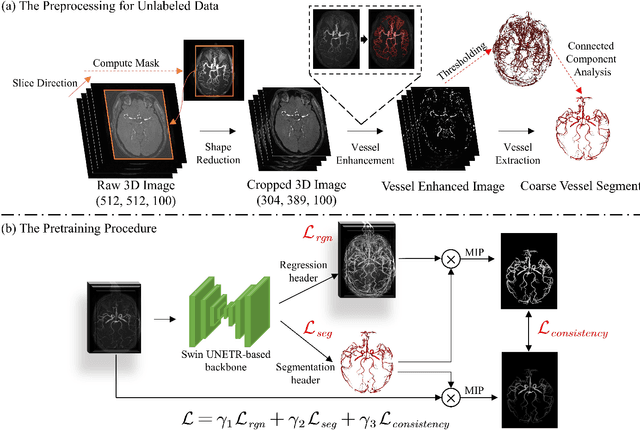
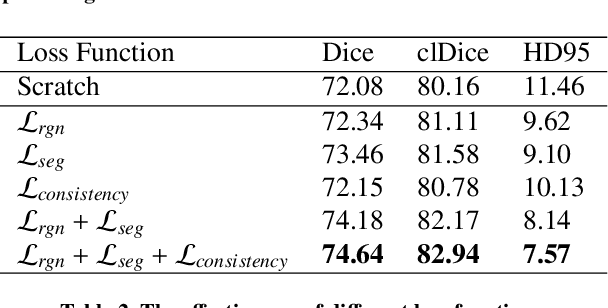
Abstract:The precise cerebrovascular segmentation in time-of-flight magnetic resonance angiography (TOF-MRA) data is crucial for clinically computer-aided diagnosis. However, the sparse distribution of cerebrovascular structures in TOF-MRA results in an exceedingly high cost for manual data labeling. The use of unlabeled TOF-MRA data holds the potential to enhance model performance significantly. In this study, we construct the largest preprocessed unlabeled TOF-MRA datasets (1510 subjects) to date. We also provide three additional labeled datasets totaling 113 subjects. Furthermore, we propose a simple yet effective pertraining strategy based on Frangi filtering, known for enhancing vessel-like structures, to fully leverage the unlabeled data for 3D cerebrovascular segmentation. Specifically, we develop a Frangi filtering-based preprocessing workflow to handle the large-scale unlabeled dataset, and a multi-task pretraining strategy is proposed to effectively utilize the preprocessed data. By employing this approach, we maximize the knowledge gained from the unlabeled data. The pretrained model is evaluated on four cerebrovascular segmentation datasets. The results have demonstrated the superior performance of our model, with an improvement of approximately 3\% compared to state-of-the-art semi- and self-supervised methods. Furthermore, the ablation studies also demonstrate the generalizability and effectiveness of the pretraining method regarding the backbone structures. The code and data have been open source at: \url{https://github.com/shigen-StoneRoot/FFPN}.
A Heterogeneous Graph Based Framework for Multimodal Neuroimaging Fusion Learning
Oct 16, 2021



Abstract:Here, we present a Heterogeneous Graph neural network for Multimodal neuroimaging fusion learning (HGM). Traditional GNN-based models usually assume the brain network is a homogeneous graph with single type of nodes and edges. However, vast literatures have shown the heterogeneity of the human brain especially between the two hemispheres. Homogeneous brain network is insufficient to model the complicated brain state. Therefore, in this work we firstly model the brain network as heterogeneous graph with multi-type nodes (i.e., left and right hemispheric nodes) and multi-type edges (i.e., intra- and inter-hemispheric edges). Besides, we also propose a self-supervised pre-training strategy based on heterogeneou brain network to address the overfitting problem due to the complex model and small sample size. Our results on two datasets show the superiority of proposed model over other multimodal methods for disease prediction task. Besides, ablation experiments show that our model with pre-training strategy can alleviate the problem of limited training sample size.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge