Franz Pfister
GLOWin: A Flow-based Invertible Generative Framework for Learning Disentangled Feature Representations in Medical Images
Mar 19, 2021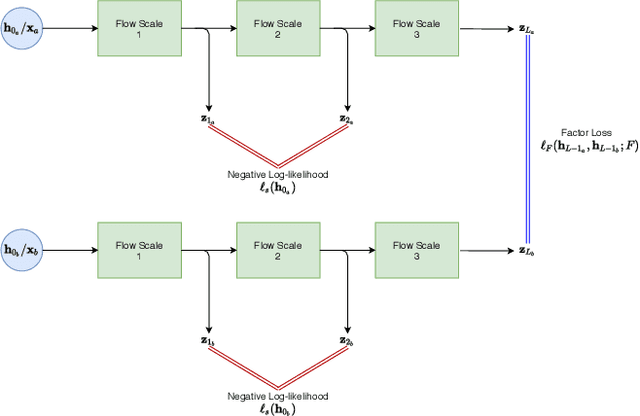


Abstract:Disentangled representations can be useful in many downstream tasks, help to make deep learning models more interpretable, and allow for control over features of synthetically generated images that can be useful in training other models that require a large number of labelled or unlabelled data. Recently, flow-based generative models have been proposed to generate realistic images by directly modeling the data distribution with invertible functions. In this work, we propose a new flow-based generative model framework, named GLOWin, that is end-to-end invertible and able to learn disentangled representations. Feature disentanglement is achieved by factorizing the latent space into components such that each component learns the representation for one generative factor. Comprehensive experiments have been conducted to evaluate the proposed method on a public brain tumor MR dataset. Quantitative and qualitative results suggest that the proposed method is effective in disentangling the features from complex medical images.
Self-Supervised Out-of-Distribution Detection in Brain CT Scans
Nov 10, 2020


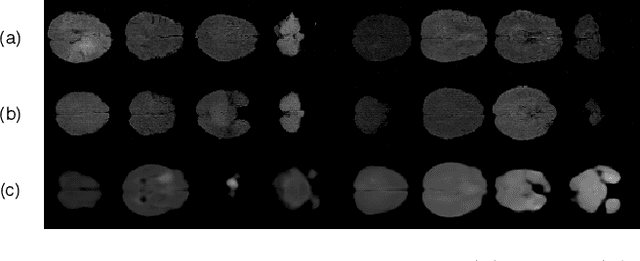
Abstract:Medical imaging data suffers from the limited availability of annotation because annotating 3D medical data is a time-consuming and expensive task. Moreover, even if the annotation is available, supervised learning-based approaches suffer highly imbalanced data. Most of the scans during the screening are from normal subjects, but there are also large variations in abnormal cases. To address these issues, recently, unsupervised deep anomaly detection methods that train the model on large-sized normal scans and detect abnormal scans by calculating reconstruction error have been reported. In this paper, we propose a novel self-supervised learning technique for anomaly detection. Our architecture largely consists of two parts: 1) Reconstruction and 2) predicting geometric transformations. By training the network to predict geometric transformations, the model could learn better image features and distribution of normal scans. In the test time, the geometric transformation predictor can assign the anomaly score by calculating the error between geometric transformation and prediction. Moreover, we further use self-supervised learning with context restoration for pretraining our model. By comparative experiments on clinical brain CT scans, the effectiveness of the proposed method has been verified.
Train, Learn, Expand, Repeat
Apr 19, 2020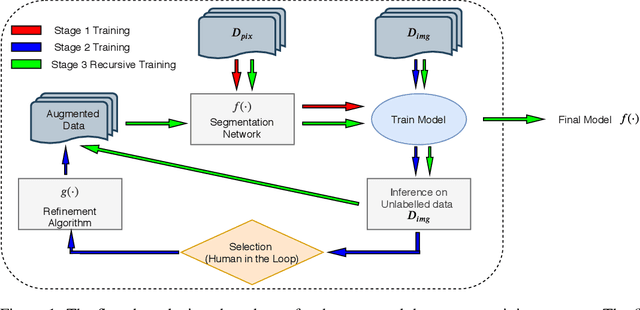
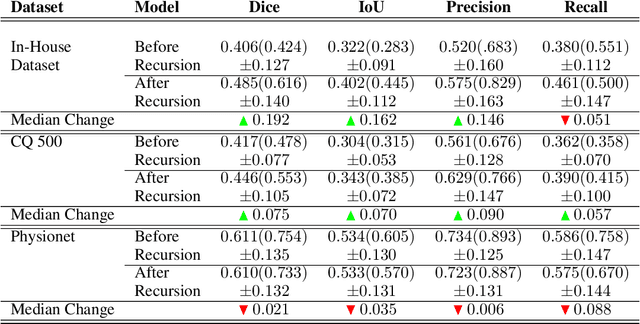
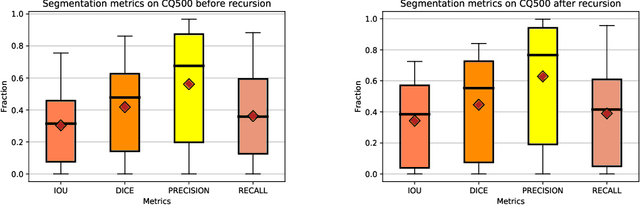
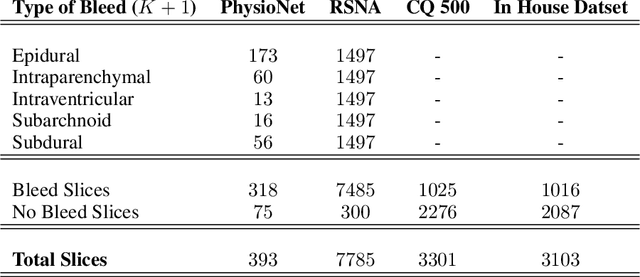
Abstract:High-quality labeled data is essential to successfully train supervised machine learning models. Although a large amount of unlabeled data is present in the medical domain, labeling poses a major challenge: medical professionals who can expertly label the data are a scarce and expensive resource. Making matters worse, voxel-wise delineation of data (e.g. for segmentation tasks) is tedious and suffers from high inter-rater variance, thus dramatically limiting available training data. We propose a recursive training strategy to perform the task of semantic segmentation given only very few training samples with pixel-level annotations. We expand on this small training set having cheaper image-level annotations using a recursive training strategy. We apply this technique on the segmentation of intracranial hemorrhage (ICH) in CT (computed tomography) scans of the brain, where typically few annotated data is available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge