Train, Learn, Expand, Repeat
Paper and Code
Apr 19, 2020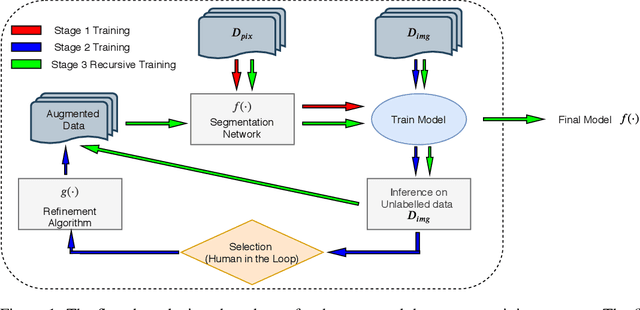
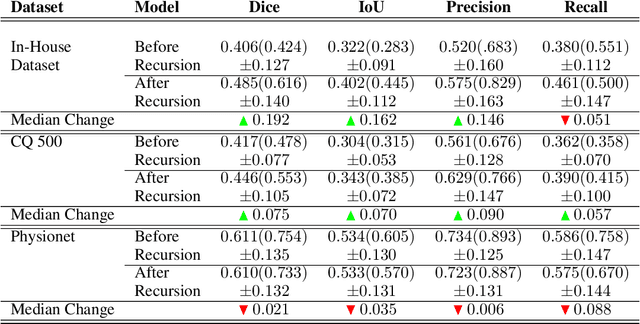
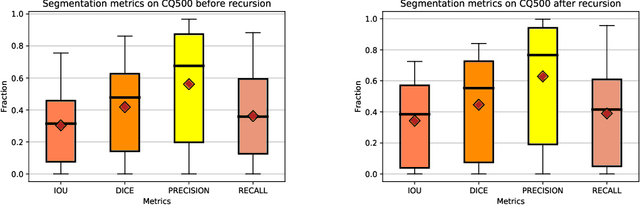
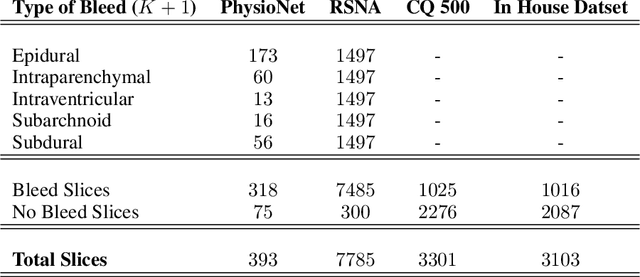
High-quality labeled data is essential to successfully train supervised machine learning models. Although a large amount of unlabeled data is present in the medical domain, labeling poses a major challenge: medical professionals who can expertly label the data are a scarce and expensive resource. Making matters worse, voxel-wise delineation of data (e.g. for segmentation tasks) is tedious and suffers from high inter-rater variance, thus dramatically limiting available training data. We propose a recursive training strategy to perform the task of semantic segmentation given only very few training samples with pixel-level annotations. We expand on this small training set having cheaper image-level annotations using a recursive training strategy. We apply this technique on the segmentation of intracranial hemorrhage (ICH) in CT (computed tomography) scans of the brain, where typically few annotated data is available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge