Francesco Paolo Casale
Random Walk Diffusion for Efficient Large-Scale Graph Generation
Aug 08, 2024Abstract:Graph generation addresses the problem of generating new graphs that have a data distribution similar to real-world graphs. While previous diffusion-based graph generation methods have shown promising results, they often struggle to scale to large graphs. In this work, we propose ARROW-Diff (AutoRegressive RandOm Walk Diffusion), a novel random walk-based diffusion approach for efficient large-scale graph generation. Our method encompasses two components in an iterative process of random walk sampling and graph pruning. We demonstrate that ARROW-Diff can scale to large graphs efficiently, surpassing other baseline methods in terms of both generation time and multiple graph statistics, reflecting the high quality of the generated graphs.
Attention-based Multi-instance Mixed Models
Nov 04, 2023



Abstract:Predicting patient features from single-cell data can unveil cellular states implicated in health and disease. Linear models and average cell type expressions are typically favored for this task for their efficiency and robustness, but they overlook the rich cell heterogeneity inherent in single-cell data. To address this gap, we introduce GMIL, a framework integrating Generalized Linear Mixed Models (GLMM) and Multiple Instance Learning (MIL), upholding the advantages of linear models while modeling cell-state heterogeneity. By leveraging predefined cell embeddings, GMIL enhances computational efficiency and aligns with recent advancements in single-cell representation learning. Our empirical results reveal that GMIL outperforms existing MIL models in single-cell datasets, uncovering new associations and elucidating biological mechanisms across different domains.
BEL: A Bag Embedding Loss for Transformer enhances Multiple Instance Whole Slide Image Classification
Mar 02, 2023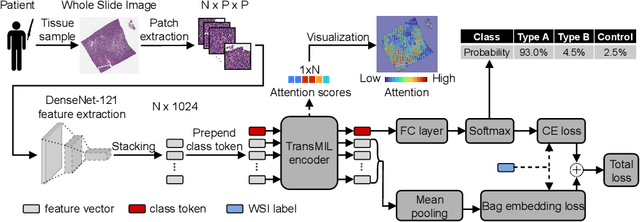


Abstract:Multiple Instance Learning (MIL) has become the predominant approach for classification tasks on gigapixel histopathology whole slide images (WSIs). Within the MIL framework, single WSIs (bags) are decomposed into patches (instances), with only WSI-level annotation available. Recent MIL approaches produce highly informative bag level representations by utilizing the transformer architecture's ability to model the dependencies between instances. However, when applied to high magnification datasets, problems emerge due to the large number of instances and the weak supervisory learning signal. To address this problem, we propose to additionally train transformers with a novel Bag Embedding Loss (BEL). BEL forces the model to learn a discriminative bag-level representation by minimizing the distance between bag embeddings of the same class and maximizing the distance between different classes. We evaluate BEL with the Transformer architecture TransMIL on two publicly available histopathology datasets, BRACS and CAMELYON17. We show that with BEL, TransMIL outperforms the baseline models on both datasets, thus contributing to the clinically highly relevant AI-based tumor classification of histological patient material.
Probabilistic Neural Architecture Search
Feb 13, 2019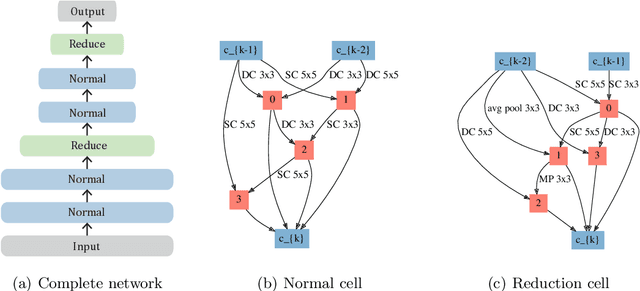

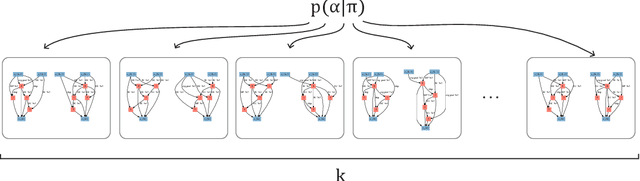

Abstract:In neural architecture search (NAS), the space of neural network architectures is automatically explored to maximize predictive accuracy for a given task. Despite the success of recent approaches, most existing methods cannot be directly applied to large scale problems because of their prohibitive computational complexity or high memory usage. In this work, we propose a Probabilistic approach to neural ARchitecture SEarCh (PARSEC) that drastically reduces memory requirements while maintaining state-of-the-art computational complexity, making it possible to directly search over more complex architectures and larger datasets. Our approach only requires as much memory as is needed to train a single architecture from our search space. This is due to a memory-efficient sampling procedure wherein we learn a probability distribution over high-performing neural network architectures. Importantly, this framework enables us to transfer the distribution of architectures learnt on smaller problems to larger ones, further reducing the computational cost. We showcase the advantages of our approach in applications to CIFAR-10 and ImageNet, where our approach outperforms methods with double its computational cost and matches the performance of methods with costs that are three orders of magnitude larger.
Gaussian Process Prior Variational Autoencoders
Oct 28, 2018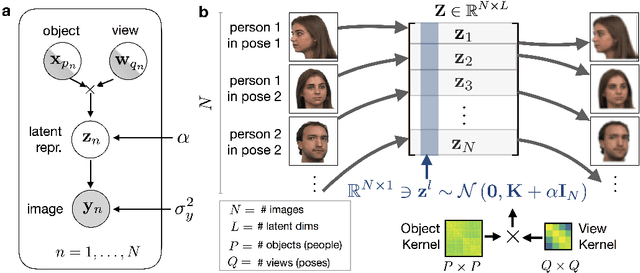
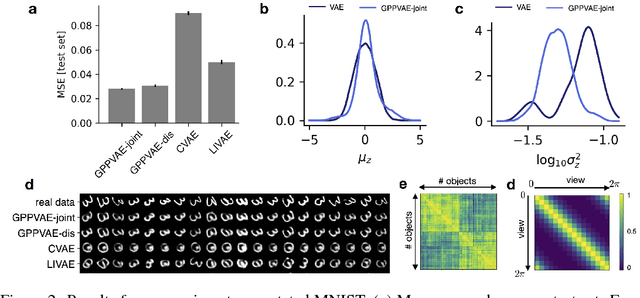
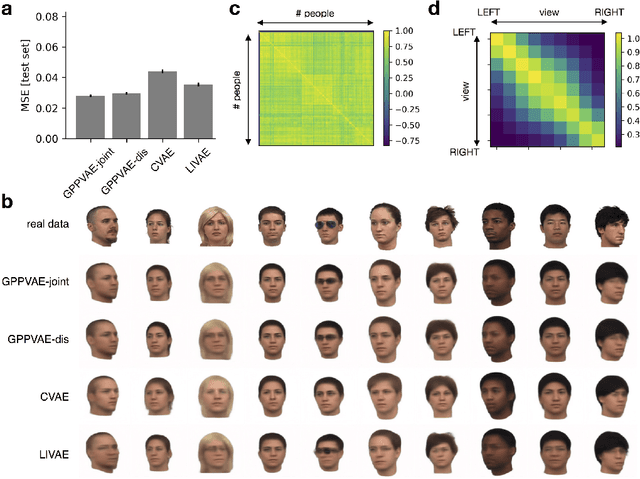
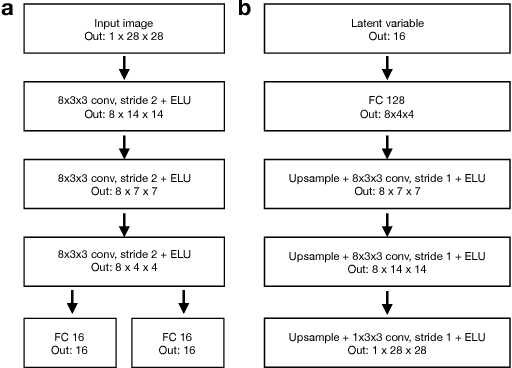
Abstract:Variational autoencoders (VAE) are a powerful and widely-used class of models to learn complex data distributions in an unsupervised fashion. One important limitation of VAEs is the prior assumption that latent sample representations are independent and identically distributed. However, for many important datasets, such as time-series of images, this assumption is too strong: accounting for covariances between samples, such as those in time, can yield to a more appropriate model specification and improve performance in downstream tasks. In this work, we introduce a new model, the Gaussian Process (GP) Prior Variational Autoencoder (GPPVAE), to specifically address this issue. The GPPVAE aims to combine the power of VAEs with the ability to model correlations afforded by GP priors. To achieve efficient inference in this new class of models, we leverage structure in the covariance matrix, and introduce a new stochastic backpropagation strategy that allows for computing stochastic gradients in a distributed and low-memory fashion. We show that our method outperforms conditional VAEs (CVAEs) and an adaptation of standard VAEs in two image data applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge