Elias Hossain
UAT-LITE: Inference-Time Uncertainty-Aware Attention for Pretrained Transformers
Feb 03, 2026Abstract:Neural NLP models are often miscalibrated, assigning high confidence to incorrect predictions, which undermines selective prediction and high-stakes deployment. Post-hoc calibration methods adjust output probabilities but leave internal computation unchanged, while ensemble and Bayesian approaches improve uncertainty at substantial training or storage cost. We propose UAT-LITE, an inference-time framework that makes self-attention uncertainty-aware using approximate Bayesian inference via Monte Carlo dropout in pretrained transformer classifiers. Token-level epistemic uncertainty is estimated from stochastic forward passes and used to modulate self-attention during contextualization, without modifying pretrained weights or training objectives. We additionally introduce a layerwise variance decomposition to diagnose how predictive uncertainty accumulates across transformer depth. Across the SQuAD 2.0 answerability, MNLI, and SST-2, UAT-LITE reduces Expected Calibration Error by approximately 20% on average relative to a fine-tuned BERT-base baseline while preserving task accuracy, and improves selective prediction and robustness under distribution shift.
Edge-Based Learning for Improved Classification Under Adversarial Noise
Apr 25, 2025Abstract:Adversarial noise introduces small perturbations in images, misleading deep learning models into misclassification and significantly impacting recognition accuracy. In this study, we analyzed the effects of Fast Gradient Sign Method (FGSM) adversarial noise on image classification and investigated whether training on specific image features can improve robustness. We hypothesize that while adversarial noise perturbs various regions of an image, edges may remain relatively stable and provide essential structural information for classification. To test this, we conducted a series of experiments using brain tumor and COVID datasets. Initially, we trained the models on clean images and then introduced subtle adversarial perturbations, which caused deep learning models to significantly misclassify the images. Retraining on a combination of clean and noisy images led to improved performance. To evaluate the robustness of the edge features, we extracted edges from the original/clean images and trained the models exclusively on edge-based representations. When noise was introduced to the images, the edge-based models demonstrated greater resilience to adversarial attacks compared to those trained on the original or clean images. These results suggest that while adversarial noise is able to exploit complex non-edge regions significantly more than edges, the improvement in the accuracy after retraining is marginally more in the original data as compared to the edges. Thus, leveraging edge-based learning can improve the resilience of deep learning models against adversarial perturbations.
Medical-GAT: Cancer Document Classification Leveraging Graph-Based Residual Network for Scenarios with Limited Data
Oct 19, 2024Abstract:Accurate classification of cancer-related medical abstracts is crucial for healthcare management and research. However, obtaining large, labeled datasets in the medical domain is challenging due to privacy concerns and the complexity of clinical data. This scarcity of annotated data impedes the development of effective machine learning models for cancer document classification. To address this challenge, we present a curated dataset of 1,874 biomedical abstracts, categorized into thyroid cancer, colon cancer, lung cancer, and generic topics. Our research focuses on leveraging this dataset to improve classification performance, particularly in data-scarce scenarios. We introduce a Residual Graph Attention Network (R-GAT) with multiple graph attention layers that capture the semantic information and structural relationships within cancer-related documents. Our R-GAT model is compared with various techniques, including transformer-based models such as Bidirectional Encoder Representations from Transformers (BERT), RoBERTa, and domain-specific models like BioBERT and Bio+ClinicalBERT. We also evaluated deep learning models (CNNs, LSTMs) and traditional machine learning models (Logistic Regression, SVM). Additionally, we explore ensemble approaches that combine deep learning models to enhance classification. Various feature extraction methods are assessed, including Term Frequency-Inverse Document Frequency (TF-IDF) with unigrams and bigrams, Word2Vec, and tokenizers from BERT and RoBERTa. The R-GAT model outperforms other techniques, achieving precision, recall, and F1 scores of 0.99, 0.97, and 0.98 for thyroid cancer; 0.96, 0.94, and 0.95 for colon cancer; 0.96, 0.99, and 0.97 for lung cancer; and 0.95, 0.96, and 0.95 for generic topics.
Learning Algorithms Made Simple
Oct 11, 2024Abstract:In this paper, we discuss learning algorithms and their importance in different types of applications which includes training to identify important patterns and features in a straightforward, easy-to-understand manner. We will review the main concepts of artificial intelligence (AI), machine learning (ML), deep learning (DL), and hybrid models. Some important subsets of Machine Learning algorithms such as supervised, unsupervised, and reinforcement learning are also discussed in this paper. These techniques can be used for some important tasks like prediction, classification, and segmentation. Convolutional Neural Networks (CNNs) are used for image and video processing and many more applications. We dive into the architecture of CNNs and how to integrate CNNs with ML algorithms to build hybrid models. This paper explores the vulnerability of learning algorithms to noise, leading to misclassification. We further discuss the integration of learning algorithms with Large Language Models (LLM) to generate coherent responses applicable to many domains such as healthcare, marketing, and finance by learning important patterns from large volumes of data. Furthermore, we discuss the next generation of learning algorithms and how we may have an unified Adaptive and Dynamic Network to perform important tasks. Overall, this article provides brief overview of learning algorithms, exploring their current state, applications and future direction.
From Questions to Insightful Answers: Building an Informed Chatbot for University Resources
May 13, 2024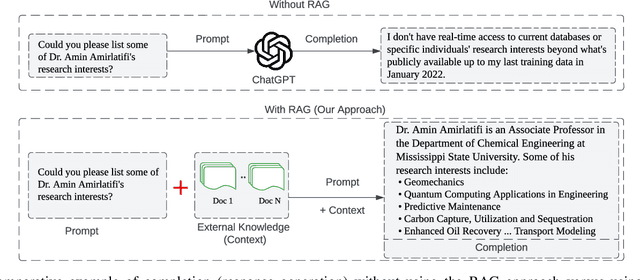
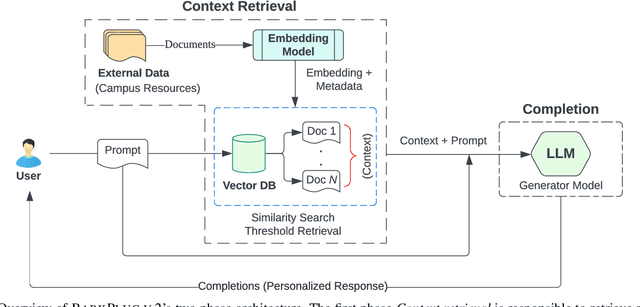
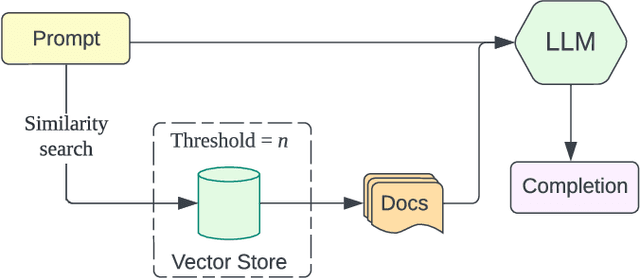
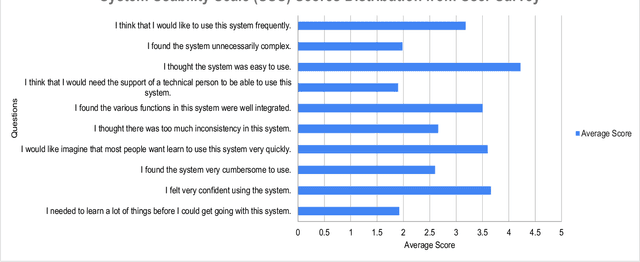
Abstract:This paper presents BARKPLUG V.2, a Large Language Model (LLM)-based chatbot system built using Retrieval Augmented Generation (RAG) pipelines to enhance the user experience and access to information within academic settings.The objective of BARKPLUG V.2 is to provide information to users about various campus resources, including academic departments, programs, campus facilities, and student resources at a university setting in an interactive fashion. Our system leverages university data as an external data corpus and ingests it into our RAG pipelines for domain-specific question-answering tasks. We evaluate the effectiveness of our system in generating accurate and pertinent responses for Mississippi State University, as a case study, using quantitative measures, employing frameworks such as Retrieval Augmented Generation Assessment(RAGAS). Furthermore, we evaluate the usability of this system via subjective satisfaction surveys using the System Usability Scale (SUS). Our system demonstrates impressive quantitative performance, with a mean RAGAS score of 0.96, and experience, as validated by usability assessments.
Natural Language Processing in Electronic Health Records in Relation to Healthcare Decision-making: A Systematic Review
Jun 22, 2023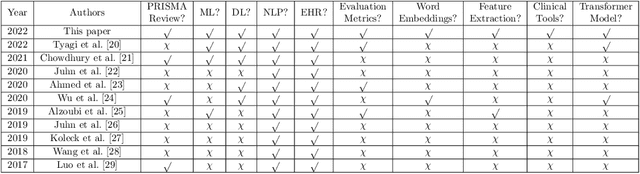
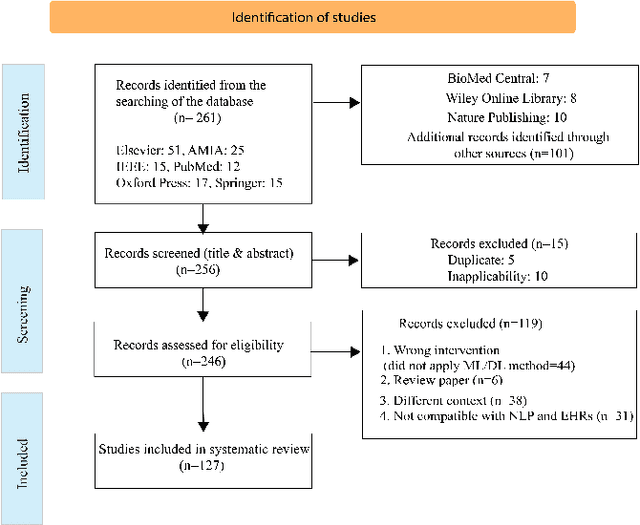
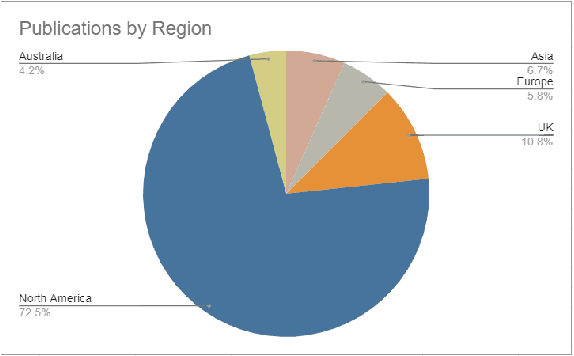
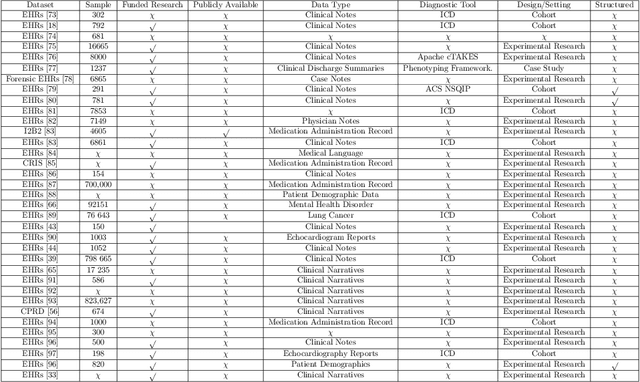
Abstract:Background: Natural Language Processing (NLP) is widely used to extract clinical insights from Electronic Health Records (EHRs). However, the lack of annotated data, automated tools, and other challenges hinder the full utilisation of NLP for EHRs. Various Machine Learning (ML), Deep Learning (DL) and NLP techniques are studied and compared to understand the limitations and opportunities in this space comprehensively. Methodology: After screening 261 articles from 11 databases, we included 127 papers for full-text review covering seven categories of articles: 1) medical note classification, 2) clinical entity recognition, 3) text summarisation, 4) deep learning (DL) and transfer learning architecture, 5) information extraction, 6) Medical language translation and 7) other NLP applications. This study follows the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. Result and Discussion: EHR was the most commonly used data type among the selected articles, and the datasets were primarily unstructured. Various ML and DL methods were used, with prediction or classification being the most common application of ML or DL. The most common use cases were: the International Classification of Diseases, Ninth Revision (ICD-9) classification, clinical note analysis, and named entity recognition (NER) for clinical descriptions and research on psychiatric disorders. Conclusion: We find that the adopted ML models were not adequately assessed. In addition, the data imbalance problem is quite important, yet we must find techniques to address this underlining problem. Future studies should address key limitations in studies, primarily identifying Lupus Nephritis, Suicide Attempts, perinatal self-harmed and ICD-9 classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge