Jeffrey Soar
Natural Language Processing in Electronic Health Records in Relation to Healthcare Decision-making: A Systematic Review
Jun 22, 2023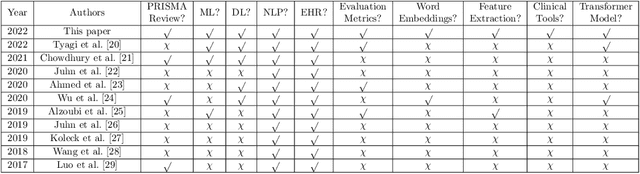
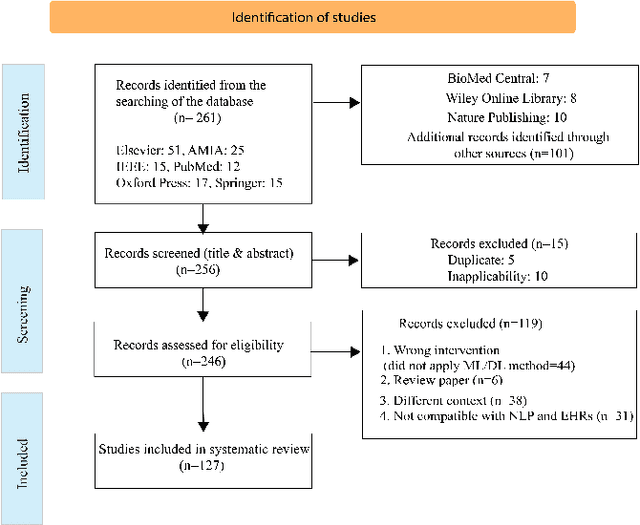
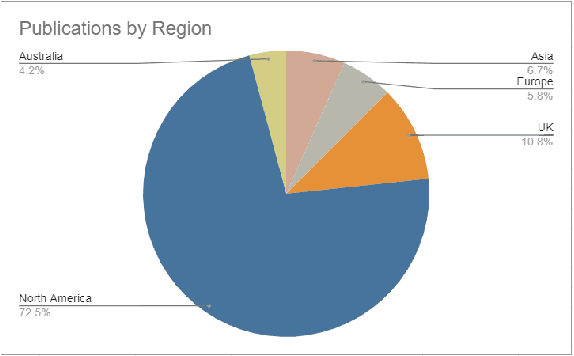
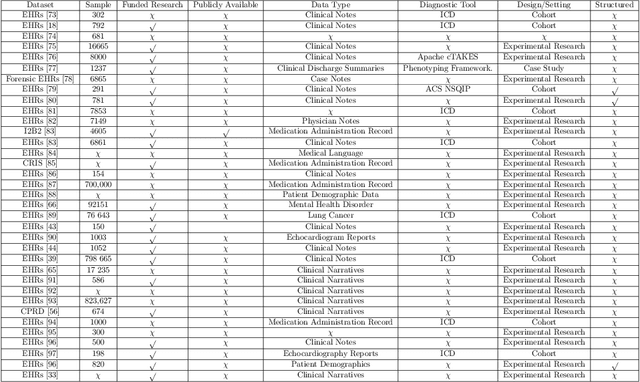
Abstract:Background: Natural Language Processing (NLP) is widely used to extract clinical insights from Electronic Health Records (EHRs). However, the lack of annotated data, automated tools, and other challenges hinder the full utilisation of NLP for EHRs. Various Machine Learning (ML), Deep Learning (DL) and NLP techniques are studied and compared to understand the limitations and opportunities in this space comprehensively. Methodology: After screening 261 articles from 11 databases, we included 127 papers for full-text review covering seven categories of articles: 1) medical note classification, 2) clinical entity recognition, 3) text summarisation, 4) deep learning (DL) and transfer learning architecture, 5) information extraction, 6) Medical language translation and 7) other NLP applications. This study follows the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. Result and Discussion: EHR was the most commonly used data type among the selected articles, and the datasets were primarily unstructured. Various ML and DL methods were used, with prediction or classification being the most common application of ML or DL. The most common use cases were: the International Classification of Diseases, Ninth Revision (ICD-9) classification, clinical note analysis, and named entity recognition (NER) for clinical descriptions and research on psychiatric disorders. Conclusion: We find that the adopted ML models were not adequately assessed. In addition, the data imbalance problem is quite important, yet we must find techniques to address this underlining problem. Future studies should address key limitations in studies, primarily identifying Lupus Nephritis, Suicide Attempts, perinatal self-harmed and ICD-9 classification.
An Overview of Ontologies and Tool Support for COVID-19 Analytics
Oct 12, 2021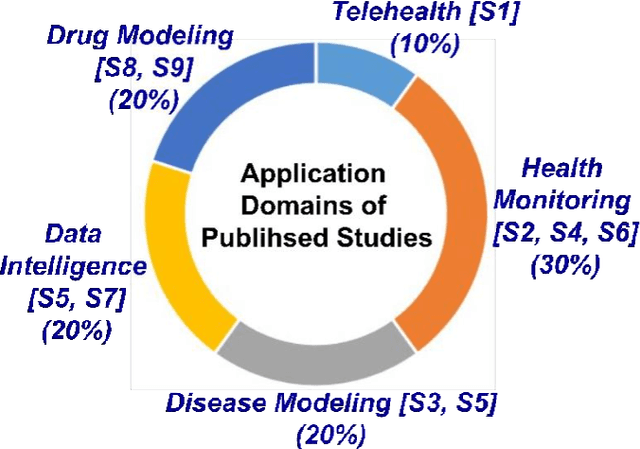
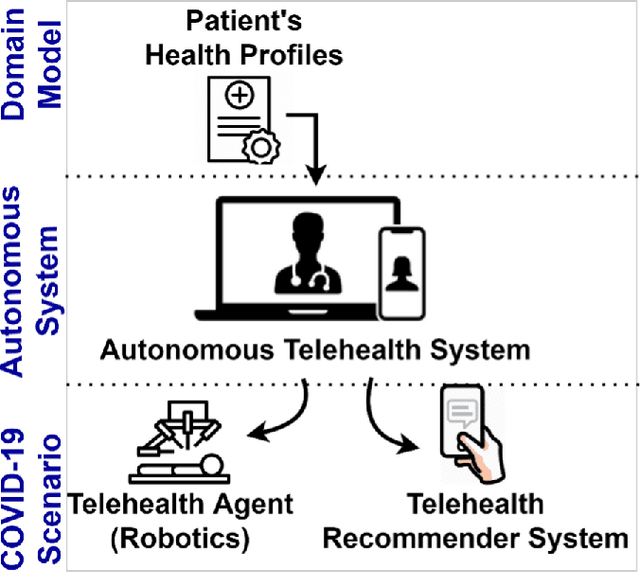
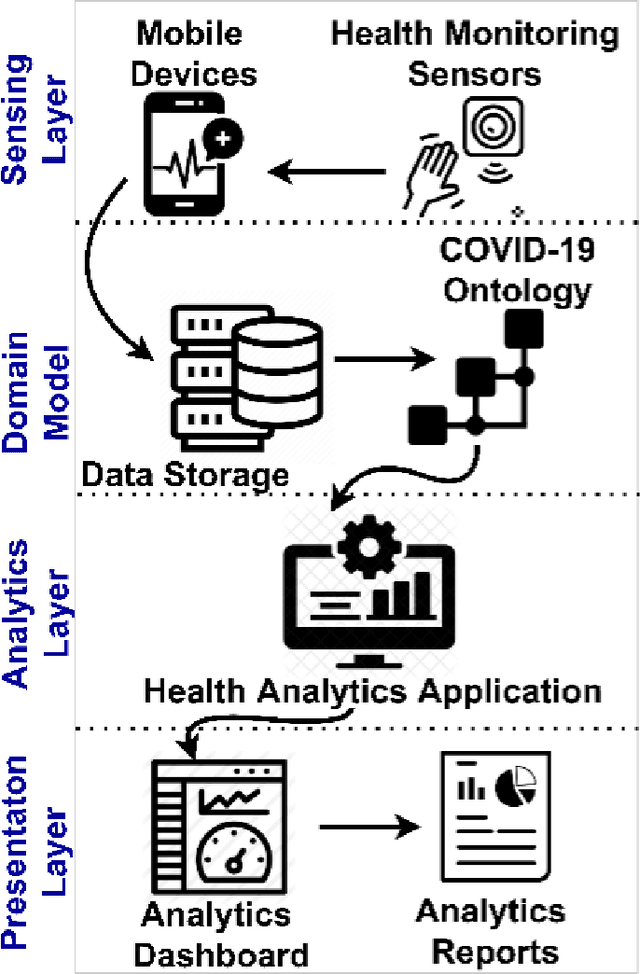
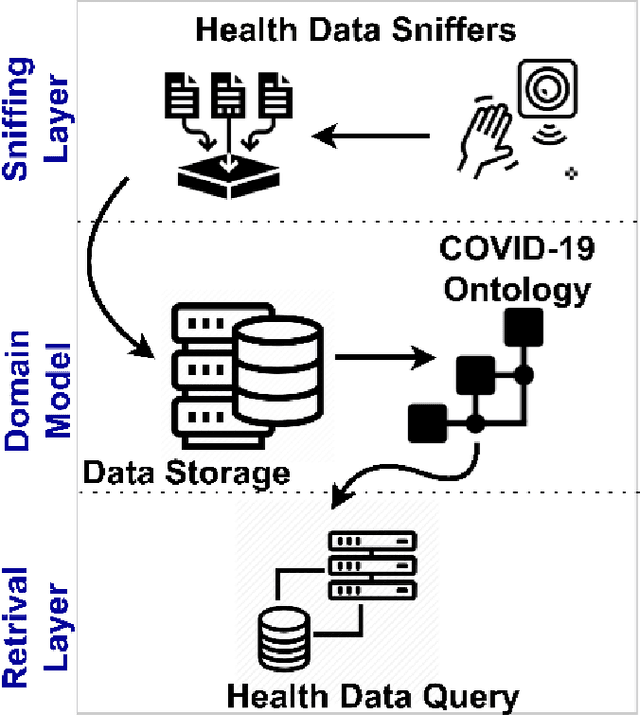
Abstract:The outbreak of the SARS-CoV-2 pandemic of the new COVID-19 disease (COVID-19 for short) demands empowering existing medical, economic, and social emergency backend systems with data analytics capabilities. An impediment in taking advantages of data analytics in these systems is the lack of a unified framework or reference model. Ontologies are highlighted as a promising solution to bridge this gap by providing a formal representation of COVID-19 concepts such as symptoms, infections rate, contact tracing, and drug modelling. Ontology-based solutions enable the integration of diverse data sources that leads to a better understanding of pandemic data, management of smart lockdowns by identifying pandemic hotspots, and knowledge-driven inference, reasoning, and recommendations to tackle surrounding issues.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge