Eleftheria Panagiotaki
Dual Deep Learning Approach for Non-invasive Renal Tumour Subtyping with VERDICT-MRI
Apr 09, 2025Abstract:This work aims to characterise renal tumour microstructure using diffusion MRI (dMRI); via the Vascular, Extracellular and Restricted Diffusion for Cytometry in Tumours (VERDICT)-MRI framework with self-supervised learning. Comprehensive datasets were acquired from 14 patients with 15 biopsy-confirmed renal tumours, with nine b-values in the range b=[0,2500]s/mm2. A three-compartment VERDICT model for renal tumours was fitted to the dMRI data using a self-supervised deep neural network, and ROIs were drawn by an experienced uroradiologist. An economical acquisition protocol for future studies with larger patient cohorts was optimised using a recursive feature selection approach. The VERDICT model described the diffusion data in renal tumours more accurately than IVIM or ADC. Combined with self-supervised deep learning, VERDICT identified significant differences in the intracellular volume fraction between cancerous and normal tissue, and in the vascular volume fraction between vascular and non-vascular. The feature selector yields a 4 b-value acquisition of b = [70,150,1000,2000], with a duration of 14 minutes.
ssVERDICT: Self-Supervised VERDICT-MRI for Enhanced Prostate Tumour Characterisation
Sep 27, 2023
Abstract:Purpose: Demonstrating and assessing self-supervised machine learning fitting of the VERDICT (Vascular, Extracellular and Restricted DIffusion for Cytometry in Tumours) model for prostate. Methods: We derive a self-supervised neural network for fitting VERDICT (ssVERDICT) that estimates parameter maps without training data. We compare the performance of ssVERDICT to two established baseline methods for fitting diffusion MRI models: conventional nonlinear least squares (NLLS) and supervised deep learning. We do this quantitatively on simulated data, by comparing the Pearson's correlation coefficient, mean-squared error (MSE), bias, and variance with respect to the simulated ground truth. We also calculate in vivo parameter maps on a cohort of 20 prostate cancer patients and compare the methods' performance in discriminating benign from cancerous tissue via Wilcoxon's signed-rank test. Results: In simulations, ssVERDICT outperforms the baseline methods (NLLS and supervised DL) in estimating all the parameters from the VERDICT prostate model in terms of Pearson's correlation coefficient, bias, and MSE. In vivo, ssVERDICT shows stronger lesion conspicuity across all parameter maps, and improves discrimination between benign and cancerous tissue over the baseline methods. Conclusion: ssVERDICT significantly outperforms state-of-the-art methods for VERDICT model fitting, and shows for the first time, fitting of a complex three-compartment biophysical model with machine learning without the requirement of explicit training labels.
Beyond Deterministic Translation for Unsupervised Domain Adaptation
Mar 11, 2022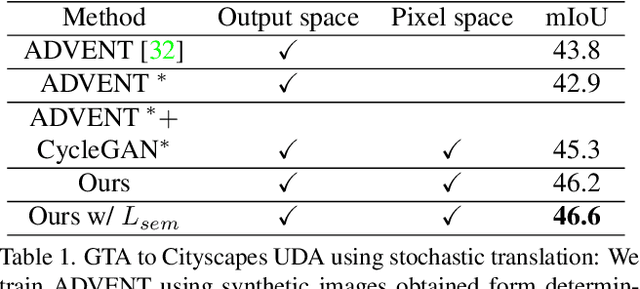

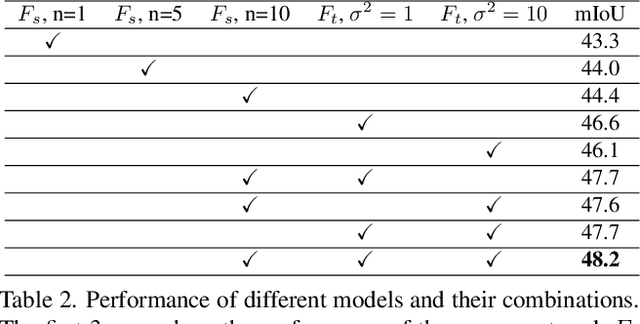

Abstract:In this work we challenge the common approach of using a one-to-one mapping ('translation') between the source and target domains in unsupervised domain adaptation (UDA). Instead, we rely on stochastic translation to capture inherent translation ambiguities. This allows us to (i) train more accurate target networks by generating multiple outputs conditioned on the same source image, leveraging both accurate translation and data augmentation for appearance variability, (ii) impute robust pseudo-labels for the target data by averaging the predictions of a source network on multiple translated versions of a single target image and (iii) train and ensemble diverse networks in the target domain by modulating the degree of stochasticity in the translations. We report improvements over strong recent baselines, leading to state-of-the-art UDA results on two challenging semantic segmentation benchmarks.
Learning to Downsample for Segmentation of Ultra-High Resolution Images
Sep 22, 2021
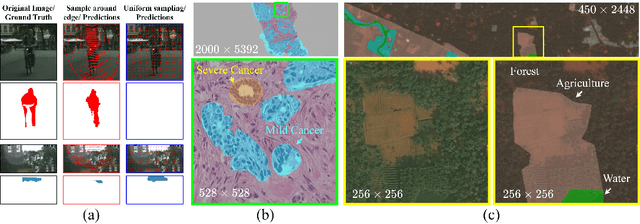
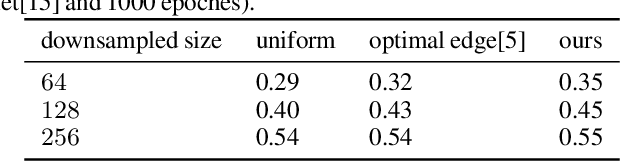
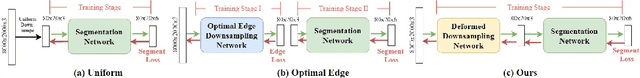
Abstract:Segmentation of ultra-high resolution images with deep learning is challenging because of their enormous size, often millions or even billions of pixels. Typical solutions drastically downsample the image uniformly to meet memory constraints, implicitly assuming all pixels equally important by sampling at the same density at all spatial locations. However this assumption is not true and compromises the performance of deep learning techniques that have proved powerful on standard-sized images. For example with uniform downsampling, see green boxed region in Fig.1, the rider and bike do not have enough corresponding samples while the trees and buildings are oversampled, and lead to a negative effect on the segmentation prediction from the low-resolution downsampled image. In this work we show that learning the spatially varying downsampling strategy jointly with segmentation offers advantages in segmenting large images with limited computational budget. Fig.1 shows that our method adapts the sampling density over different locations so that more samples are collected from the small important regions and less from the others, which in turn leads to better segmentation accuracy. We show on two public and one local high-resolution datasets that our method consistently learns sampling locations preserving more information and boosting segmentation accuracy over baseline methods.
Unsupervised Domain Adaptation with Semantic Consistency across Heterogeneous Modalities for MRI Prostate Lesion Segmentation
Sep 19, 2021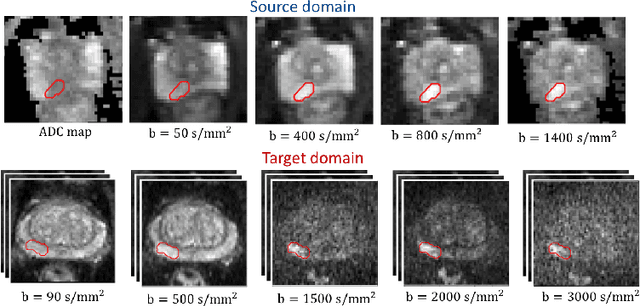

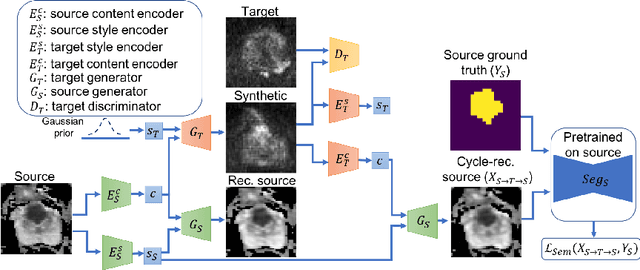
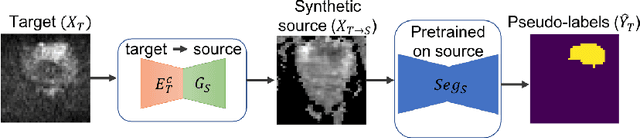
Abstract:Any novel medical imaging modality that differs from previous protocols e.g. in the number of imaging channels, introduces a new domain that is heterogeneous from previous ones. This common medical imaging scenario is rarely considered in the domain adaptation literature, which handles shifts across domains of the same dimensionality. In our work we rely on stochastic generative modeling to translate across two heterogeneous domains at pixel space and introduce two new loss functions that promote semantic consistency. Firstly, we introduce a semantic cycle-consistency loss in the source domain to ensure that the translation preserves the semantics. Secondly, we introduce a pseudo-labelling loss, where we translate target data to source, label them by a source-domain network, and use the generated pseudo-labels to supervise the target-domain network. Our results show that this allows us to extract systematically better representations for the target domain. In particular, we address the challenge of enhancing performance on VERDICT-MRI, an advanced diffusion-weighted imaging technique, by exploiting labeled mp-MRI data. When compared to several unsupervised domain adaptation approaches, our approach yields substantial improvements, that consistently carry over to the semi-supervised and supervised learning settings.
Harnessing Uncertainty in Domain Adaptation for MRI Prostate Lesion Segmentation
Oct 14, 2020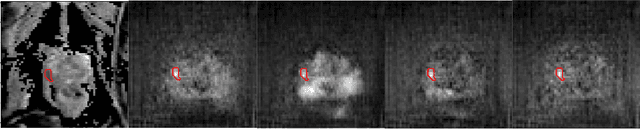
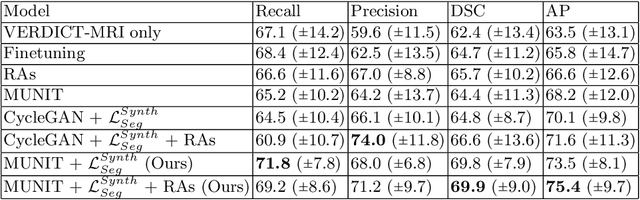
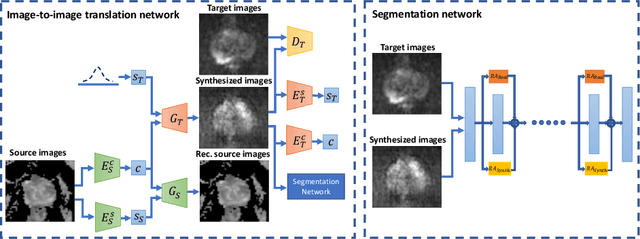
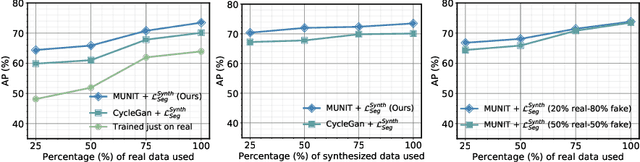
Abstract:The need for training data can impede the adoption of novel imaging modalities for learning-based medical image analysis. Domain adaptation methods partially mitigate this problem by translating training data from a related source domain to a novel target domain, but typically assume that a one-to-one translation is possible. Our work addresses the challenge of adapting to a more informative target domain where multiple target samples can emerge from a single source sample. In particular we consider translating from mp-MRI to VERDICT, a richer MRI modality involving an optimized acquisition protocol for cancer characterization. We explicitly account for the inherent uncertainty of this mapping and exploit it to generate multiple outputs conditioned on a single input. Our results show that this allows us to extract systematically better image representations for the target domain, when used in tandem with both simple, CycleGAN-based baselines, as well as more powerful approaches that integrate discriminative segmentation losses and/or residual adapters. When compared to its deterministic counterparts, our approach yields substantial improvements across a broad range of dataset sizes, increasingly strong baselines, and evaluation measures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge