Egbert Castro
ProtSCAPE: Mapping the landscape of protein conformations in molecular dynamics
Oct 27, 2024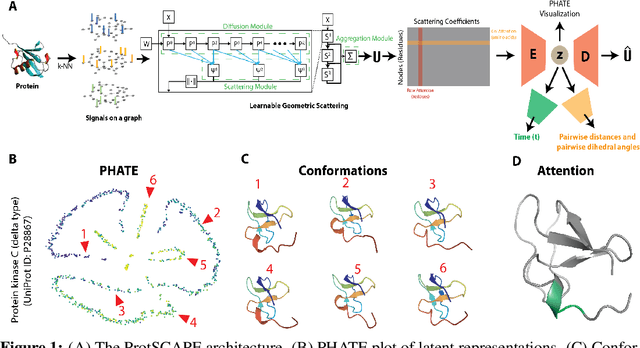
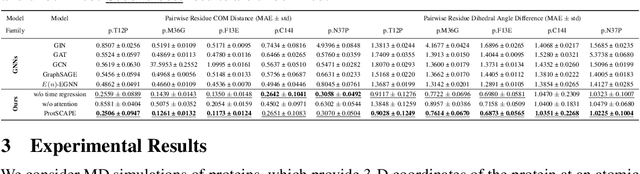
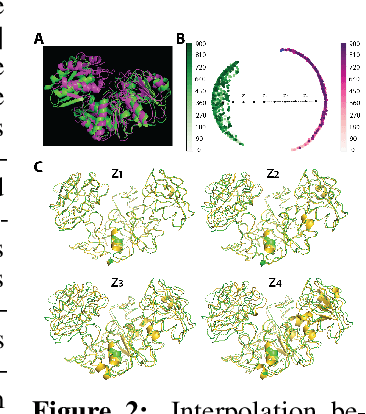
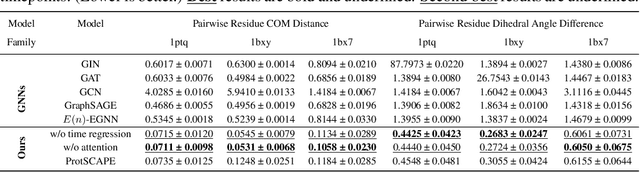
Abstract:Understanding the dynamic nature of protein structures is essential for comprehending their biological functions. While significant progress has been made in predicting static folded structures, modeling protein motions on microsecond to millisecond scales remains challenging. To address these challenges, we introduce a novel deep learning architecture, Protein Transformer with Scattering, Attention, and Positional Embedding (ProtSCAPE), which leverages the geometric scattering transform alongside transformer-based attention mechanisms to capture protein dynamics from molecular dynamics (MD) simulations. ProtSCAPE utilizes the multi-scale nature of the geometric scattering transform to extract features from protein structures conceptualized as graphs and integrates these features with dual attention structures that focus on residues and amino acid signals, generating latent representations of protein trajectories. Furthermore, ProtSCAPE incorporates a regression head to enforce temporally coherent latent representations.
Guided Generative Protein Design using Regularized Transformers
Jan 24, 2022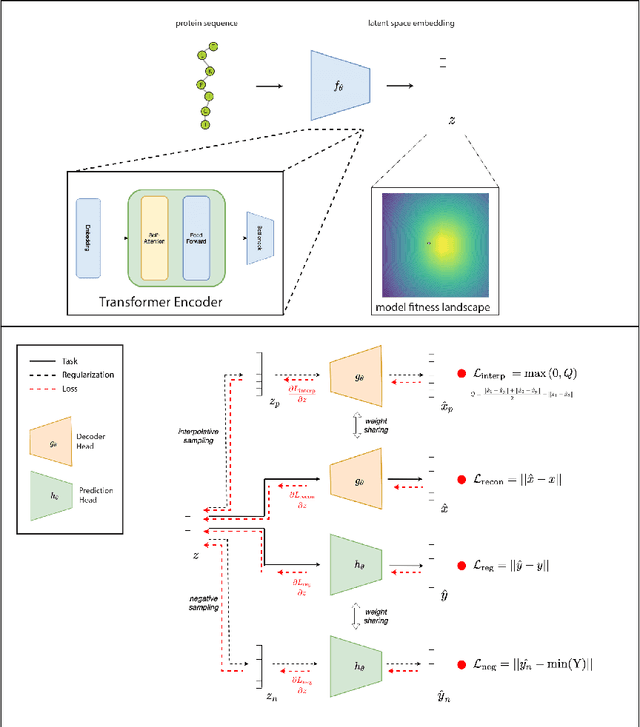
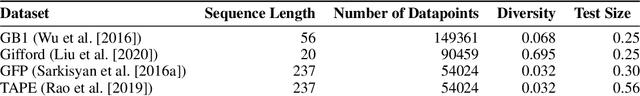
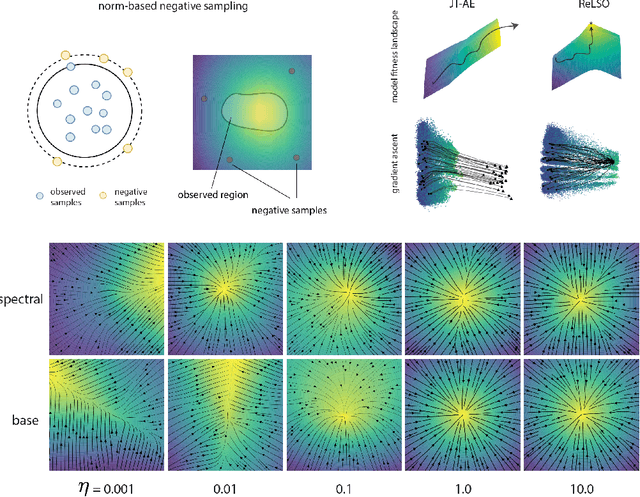
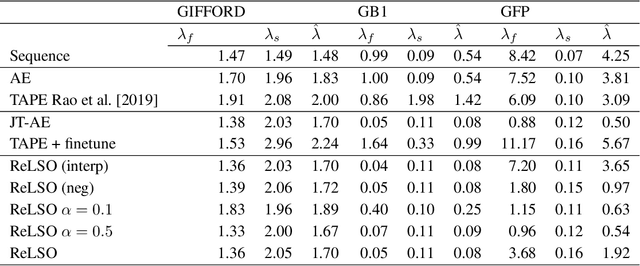
Abstract:The development of powerful natural language models have increased the ability to learn meaningful representations of protein sequences. In addition, advances in high-throughput mutagenesis, directed evolution, and next-generation sequencing have allowed for the accumulation of large amounts of labeled fitness data. Leveraging these two trends, we introduce Regularized Latent Space Optimization (ReLSO), a deep transformer-based autoencoder which is trained to jointly generate sequences as well as predict fitness. Using ReLSO, we explicitly model the underlying sequence-function landscape of large labeled datasets and optimize within latent space using gradient-based methods. Through regularized prediction heads, ReLSO introduces a powerful protein sequence encoder and novel approach for efficient fitness landscape traversal.
Uncovering the Folding Landscape of RNA Secondary Structure with Deep Graph Embeddings
Jun 16, 2020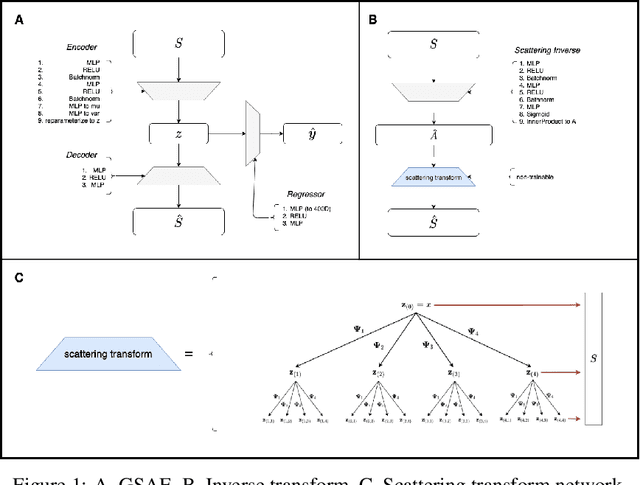
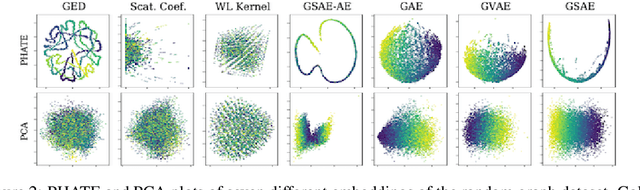
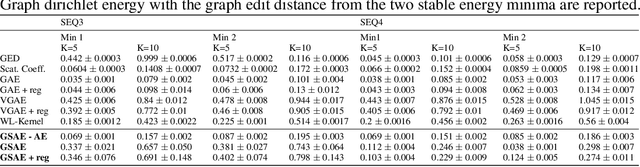
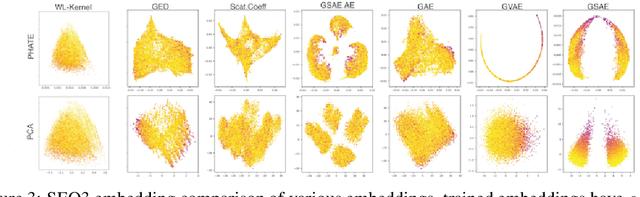
Abstract:Biomolecular graph analysis has recently gained much attention in the emerging field of geometric deep learning. While numerous approaches aim to train classifiers that accurately predict molecular properties from graphs that encode their structure, an equally important task is to organize biomolecular graphs in ways that expose meaningful relations and variations between them. We propose a geometric scattering autoencoder (GSAE) network for learning such graph embeddings. Our embedding network first extracts rich graph features using the recently proposed geometric scattering transform. Then, it leverages a semi-supervised variational autoencoder to extract a low-dimensional embedding that retains the information in these features that enable prediction of molecular properties as well as characterize graphs. Our approach is based on the intuition that geometric scattering generates multi-resolution features with in-built invariance to deformations, but as they are unsupervised, these features may not be tuned for optimally capturing relevant domain-specific properties. We demonstrate the effectiveness of our approach to data exploration of RNA foldings. Like proteins, RNA molecules can fold to create low energy functional structures such as hairpins, but the landscape of possible folds and fold sequences are not well visualized by existing methods. We show that GSAE organizes RNA graphs both by structure and energy, accurately reflecting bistable RNA structures. Furthermore, it enables interpolation of embedded molecule sequences mimicking folding trajectories. Finally, using an auxiliary inverse-scattering model, we demonstrate our ability to generate synthetic RNA graphs along the trajectory thus providing hypothetical folding sequences for further analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge