Abhinav Godavarthi
Guided Generative Protein Design using Regularized Transformers
Jan 24, 2022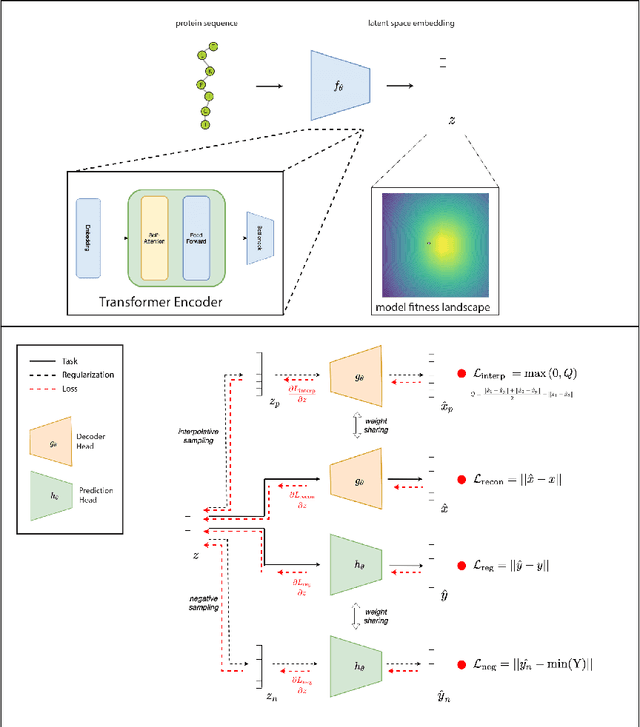
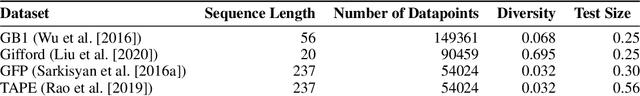
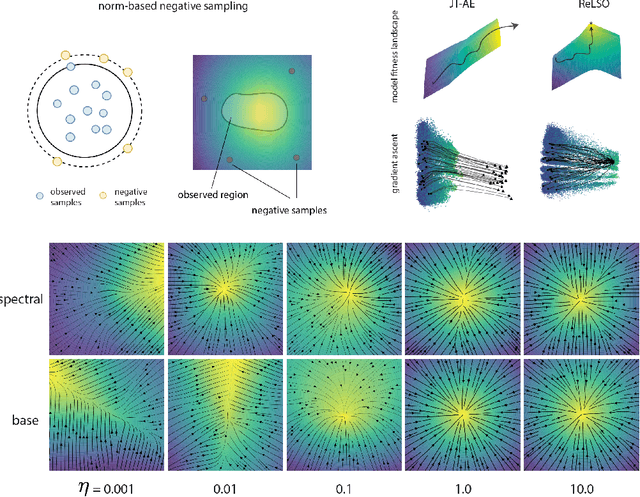
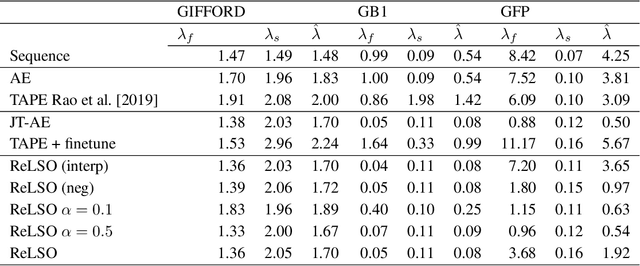
Abstract:The development of powerful natural language models have increased the ability to learn meaningful representations of protein sequences. In addition, advances in high-throughput mutagenesis, directed evolution, and next-generation sequencing have allowed for the accumulation of large amounts of labeled fitness data. Leveraging these two trends, we introduce Regularized Latent Space Optimization (ReLSO), a deep transformer-based autoencoder which is trained to jointly generate sequences as well as predict fitness. Using ReLSO, we explicitly model the underlying sequence-function landscape of large labeled datasets and optimize within latent space using gradient-based methods. Through regularized prediction heads, ReLSO introduces a powerful protein sequence encoder and novel approach for efficient fitness landscape traversal.
Multimodal data visualization, denoising and clustering with integrated diffusion
Feb 12, 2021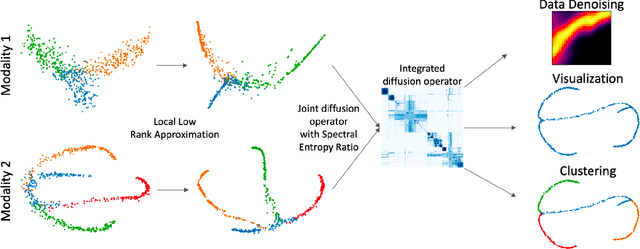
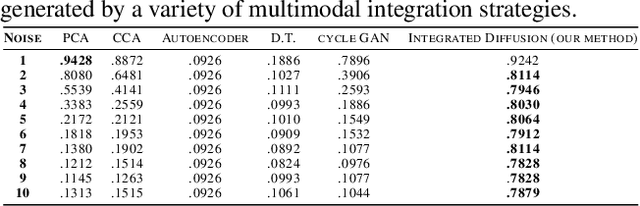
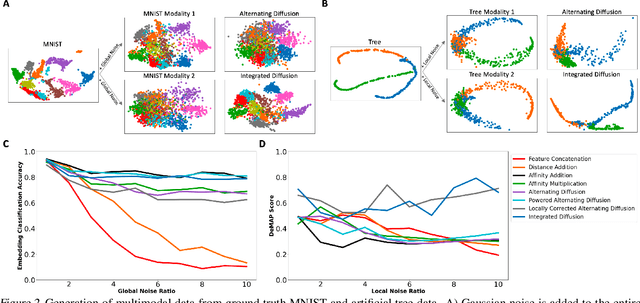
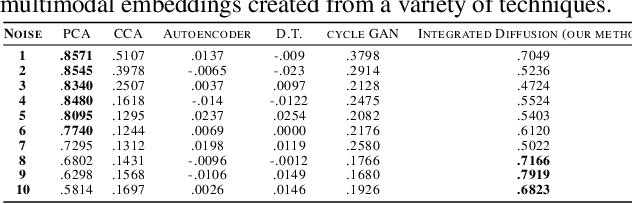
Abstract:We propose a method called integrated diffusion for combining multimodal datasets, or data gathered via several different measurements on the same system, to create a joint data diffusion operator. As real world data suffers from both local and global noise, we introduce mechanisms to optimally calculate a diffusion operator that reflects the combined information from both modalities. We show the utility of this joint operator in data denoising, visualization and clustering, performing better than other methods to integrate and analyze multimodal data. We apply our method to multi-omic data generated from blood cells, measuring both gene expression and chromatin accessibility. Our approach better visualizes the geometry of the joint data, captures known cross-modality associations and identifies known cellular populations. More generally, integrated diffusion is broadly applicable to multimodal datasets generated in many medical and biological systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge