De-Jun Fan
Annotation-Efficient Polyp Segmentation via Active Learning
Mar 21, 2024Abstract:Deep learning-based techniques have proven effective in polyp segmentation tasks when provided with sufficient pixel-wise labeled data. However, the high cost of manual annotation has created a bottleneck for model generalization. To minimize annotation costs, we propose a deep active learning framework for annotation-efficient polyp segmentation. In practice, we measure the uncertainty of each sample by examining the similarity between features masked by the prediction map of the polyp and the background area. Since the segmentation model tends to perform weak in samples with indistinguishable features of foreground and background areas, uncertainty sampling facilitates the fitting of under-learning data. Furthermore, clustering image-level features weighted by uncertainty identify samples that are both uncertain and representative. To enhance the selectivity of the active selection strategy, we propose a novel unsupervised feature discrepancy learning mechanism. The selection strategy and feature optimization work in tandem to achieve optimal performance with a limited annotation budget. Extensive experimental results have demonstrated that our proposed method achieved state-of-the-art performance compared to other competitors on both a public dataset and a large-scale in-house dataset.
Lesion-aware Dynamic Kernel for Polyp Segmentation
Jan 12, 2023Abstract:Automatic and accurate polyp segmentation plays an essential role in early colorectal cancer diagnosis. However, it has always been a challenging task due to 1) the diverse shape, size, brightness and other appearance characteristics of polyps, 2) the tiny contrast between concealed polyps and their surrounding regions. To address these problems, we propose a lesion-aware dynamic network (LDNet) for polyp segmentation, which is a traditional u-shape encoder-decoder structure incorporated with a dynamic kernel generation and updating scheme. Specifically, the designed segmentation head is conditioned on the global context features of the input image and iteratively updated by the extracted lesion features according to polyp segmentation predictions. This simple but effective scheme endows our model with powerful segmentation performance and generalization capability. Besides, we utilize the extracted lesion representation to enhance the feature contrast between the polyp and background regions by a tailored lesion-aware cross-attention module (LCA), and design an efficient self-attention module (ESA) to capture long-range context relations, further improving the segmentation accuracy. Extensive experiments on four public polyp benchmarks and our collected large-scale polyp dataset demonstrate the superior performance of our method compared with other state-of-the-art approaches. The source code is available at https://github.com/ReaFly/LDNet.
Cross-level Contrastive Learning and Consistency Constraint for Semi-supervised Medical Image Segmentation
Feb 13, 2022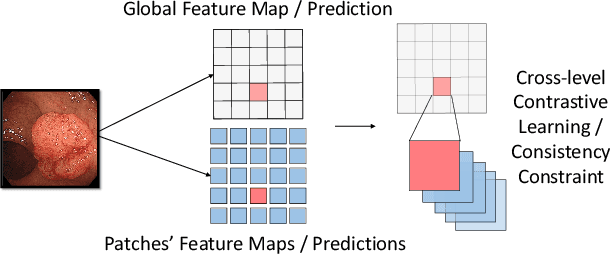
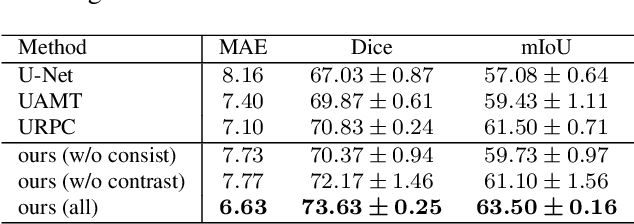
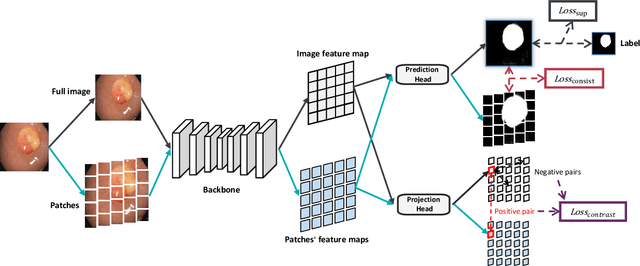
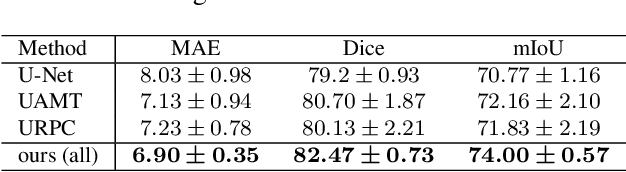
Abstract:Semi-supervised learning (SSL), which aims at leveraging a few labeled images and a large number of unlabeled images for network training, is beneficial for relieving the burden of data annotation in medical image segmentation. According to the experience of medical imaging experts, local attributes such as texture, luster and smoothness are very important factors for identifying target objects like lesions and polyps in medical images. Motivated by this, we propose a cross-level contrastive learning scheme to enhance representation capacity for local features in semi-supervised medical image segmentation. Compared to existing image-wise, patch-wise and point-wise contrastive learning algorithms, our devised method is capable of exploring more complex similarity cues, namely the relational characteristics between global and local patch-wise representations. Additionally, for fully making use of cross-level semantic relations, we devise a novel consistency constraint that compares the predictions of patches against those of the full image. With the help of the cross-level contrastive learning and consistency constraint, the unlabelled data can be effectively explored to improve segmentation performance on two medical image datasets for polyp and skin lesion segmentation respectively. Code of our approach is available.
Deep Transformers for Fast Small Intestine Grounding in Capsule Endoscope Video
Apr 07, 2021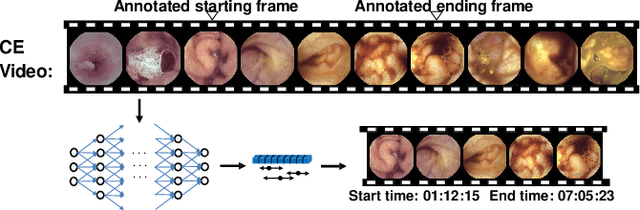
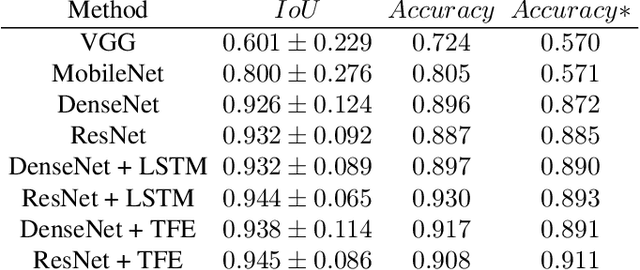
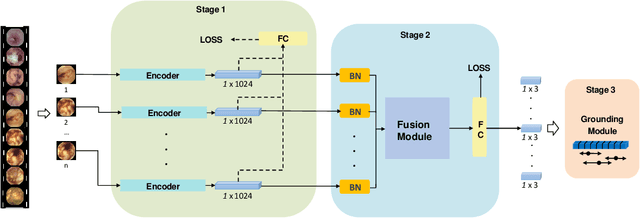
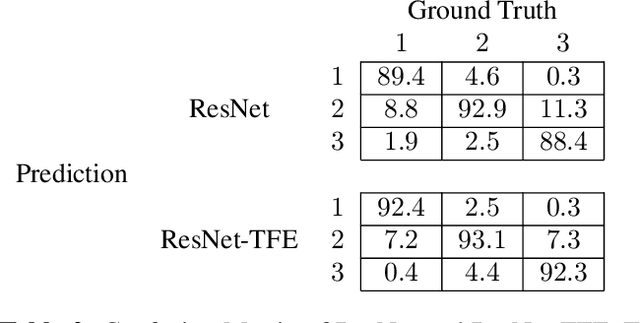
Abstract:Capsule endoscopy is an evolutional technique for examining and diagnosing intractable gastrointestinal diseases. Because of the huge amount of data, analyzing capsule endoscope videos is very time-consuming and labor-intensive for gastrointestinal medicalists. The development of intelligent long video analysis algorithms for regional positioning and analysis of capsule endoscopic video is therefore essential to reduce the workload of clinicians and assist in improving the accuracy of disease diagnosis. In this paper, we propose a deep model to ground shooting range of small intestine from a capsule endoscope video which has duration of tens of hours. This is the first attempt to attack the small intestine grounding task using deep neural network method. We model the task as a 3-way classification problem, in which every video frame is categorized into esophagus/stomach, small intestine or colorectum. To explore long-range temporal dependency, a transformer module is built to fuse features of multiple neighboring frames. Based on the classification model, we devise an efficient search algorithm to efficiently locate the starting and ending shooting boundaries of the small intestine. Without searching the small intestine exhaustively in the full video, our method is implemented via iteratively separating the video segment along the direction to the target boundary in the middle. We collect 113 videos from a local hospital to validate our method. In the 5-fold cross validation, the average IoU between the small intestine segments located by our method and the ground-truths annotated by broad-certificated gastroenterologists reaches 0.945.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge