David Elizondo
A Three-Level Alignment Framework for Large-Scale 3D Retrieval and Controlled 4D Generation
Dec 25, 2025Abstract:We introduce Uni4D, a unified framework for large scale open vocabulary 3D retrieval and controlled 4D generation based on structured three level alignment across text, 3D models, and image modalities. Built upon the Align3D 130 dataset, Uni4D employs a 3D text multi head attention and search model to optimize text to 3D retrieval through improved semantic alignment. The framework further strengthens cross modal alignment through three components: precise text to 3D retrieval, multi view 3D to image alignment, and image to text alignment for generating temporally consistent 4D assets. Experimental results demonstrate that Uni4D achieves high quality 3D retrieval and controllable 4D generation, advancing dynamic multimodal understanding and practical applications.
Learning Dynamic Scene Reconstruction with Sinusoidal Geometric Priors
Dec 25, 2025Abstract:We propose SirenPose, a novel loss function that combines the periodic activation properties of sinusoidal representation networks with geometric priors derived from keypoint structures to improve the accuracy of dynamic 3D scene reconstruction. Existing approaches often struggle to maintain motion modeling accuracy and spatiotemporal consistency in fast moving and multi target scenes. By introducing physics inspired constraint mechanisms, SirenPose enforces coherent keypoint predictions across both spatial and temporal dimensions. We further expand the training dataset to 600,000 annotated instances to support robust learning. Experimental results demonstrate that models trained with SirenPose achieve significant improvements in spatiotemporal consistency metrics compared to prior methods, showing superior performance in handling rapid motion and complex scene changes.
mldr.resampling: Efficient Reference Implementations of Multilabel Resampling Algorithms
May 30, 2023Abstract:Resampling algorithms are a useful approach to deal with imbalanced learning in multilabel scenarios. These methods have to deal with singularities in the multilabel data, such as the occurrence of frequent and infrequent labels in the same instance. Implementations of these methods are sometimes limited to the pseudocode provided by their authors in a paper. This Original Software Publication presents mldr.resampling, a software package that provides reference implementations for eleven multilabel resampling methods, with an emphasis on efficiency since these algorithms are usually time-consuming.
Semi-supervised Deep Learning for Image Classification with Distribution Mismatch: A Survey
Mar 10, 2022
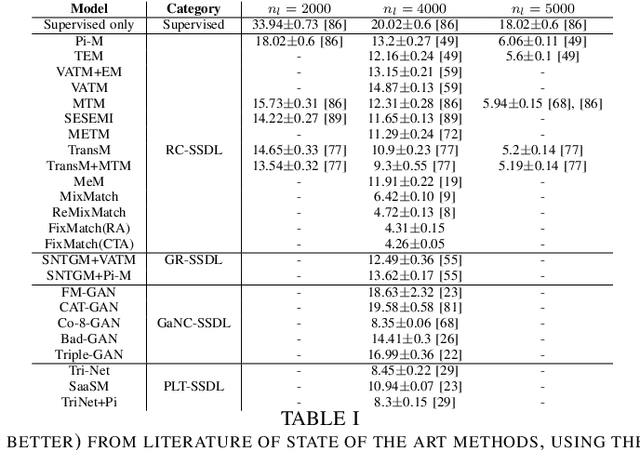
Abstract:Deep learning methodologies have been employed in several different fields, with an outstanding success in image recognition applications, such as material quality control, medical imaging, autonomous driving, etc. Deep learning models rely on the abundance of labelled observations to train a prospective model. These models are composed of millions of parameters to estimate, increasing the need of more training observations. Frequently it is expensive to gather labelled observations of data, making the usage of deep learning models not ideal, as the model might over-fit data. In a semi-supervised setting, unlabelled data is used to improve the levels of accuracy and generalization of a model with small labelled datasets. Nevertheless, in many situations different unlabelled data sources might be available. This raises the risk of a significant distribution mismatch between the labelled and unlabelled datasets. Such phenomena can cause a considerable performance hit to typical semi-supervised deep learning frameworks, which often assume that both labelled and unlabelled datasets are drawn from similar distributions. Therefore, in this paper we study the latest approaches for semi-supervised deep learning for image recognition. Emphasis is made in semi-supervised deep learning models designed to deal with a distribution mismatch between the labelled and unlabelled datasets. We address open challenges with the aim to encourage the community to tackle them, and overcome the high data demand of traditional deep learning pipelines under real-world usage settings.
Dealing with Distribution Mismatch in Semi-supervised Deep Learning for Covid-19 Detection Using Chest X-ray Images: A Novel Approach Using Feature Densities
Aug 17, 2021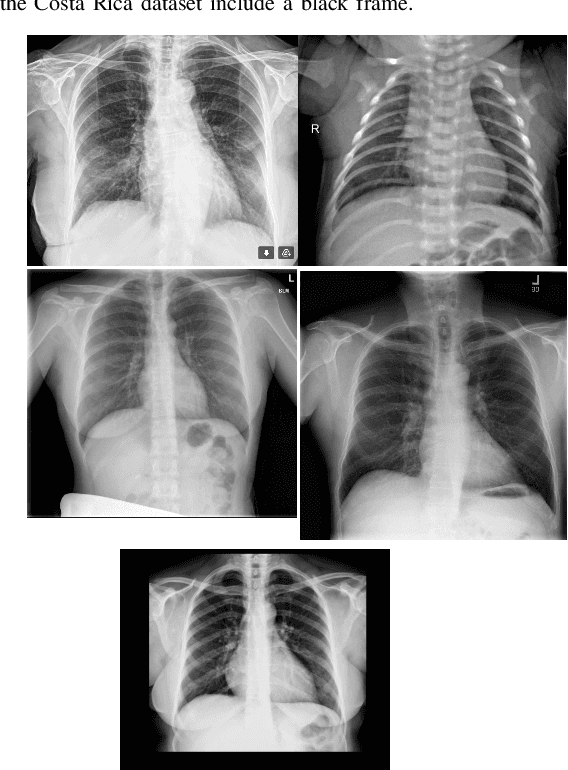

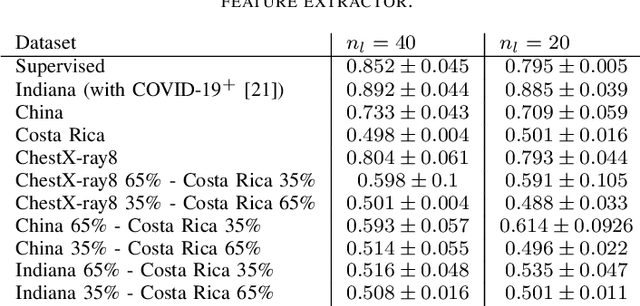
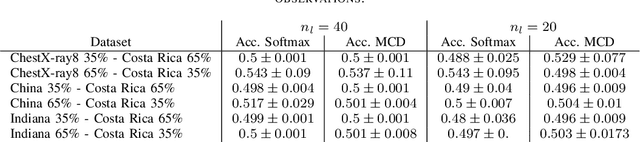
Abstract:In the context of the global coronavirus pandemic, different deep learning solutions for infected subject detection using chest X-ray images have been proposed. However, deep learning models usually need large labelled datasets to be effective. Semi-supervised deep learning is an attractive alternative, where unlabelled data is leveraged to improve the overall model's accuracy. However, in real-world usage settings, an unlabelled dataset might present a different distribution than the labelled dataset (i.e. the labelled dataset was sampled from a target clinic and the unlabelled dataset from a source clinic). This results in a distribution mismatch between the unlabelled and labelled datasets. In this work, we assess the impact of the distribution mismatch between the labelled and the unlabelled datasets, for a semi-supervised model trained with chest X-ray images, for COVID-19 detection. Under strong distribution mismatch conditions, we found an accuracy hit of almost 30\%, suggesting that the unlabelled dataset distribution has a strong influence in the behaviour of the model. Therefore, we propose a straightforward approach to diminish the impact of such distribution mismatch. Our proposed method uses a density approximation of the feature space. It is built upon the target dataset to filter out the observations in the source unlabelled dataset that might harm the accuracy of the semi-supervised model. It assumes that a small labelled source dataset is available together with a larger source unlabelled dataset. Our proposed method does not require any model training, it is simple and computationally cheap. We compare our proposed method against two popular state of the art out-of-distribution data detectors, which are also cheap and simple to implement. In our tests, our method yielded accuracy gains of up to 32\%, when compared to the previous state of the art methods.
A Real Use Case of Semi-Supervised Learning for Mammogram Classification in a Local Clinic of Costa Rica
Jul 24, 2021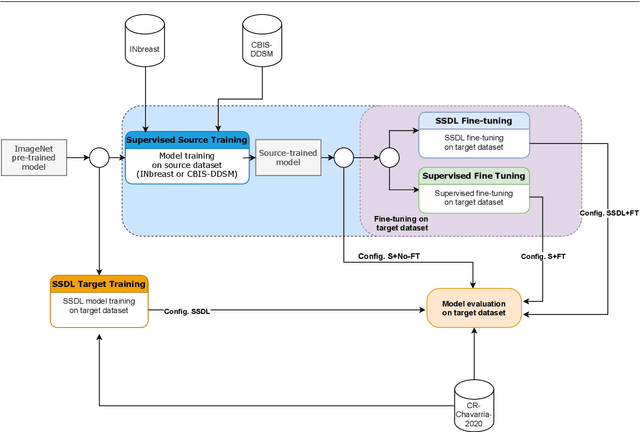
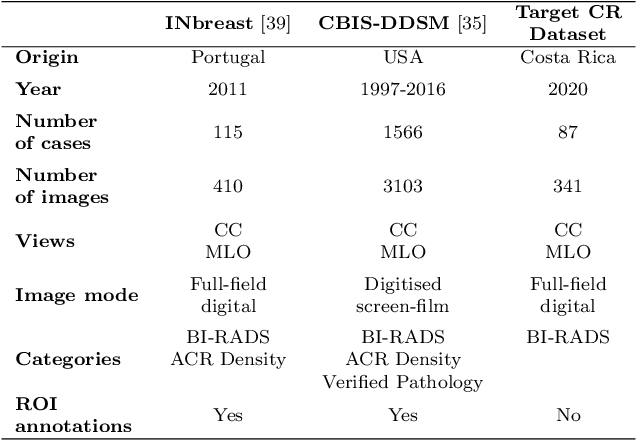
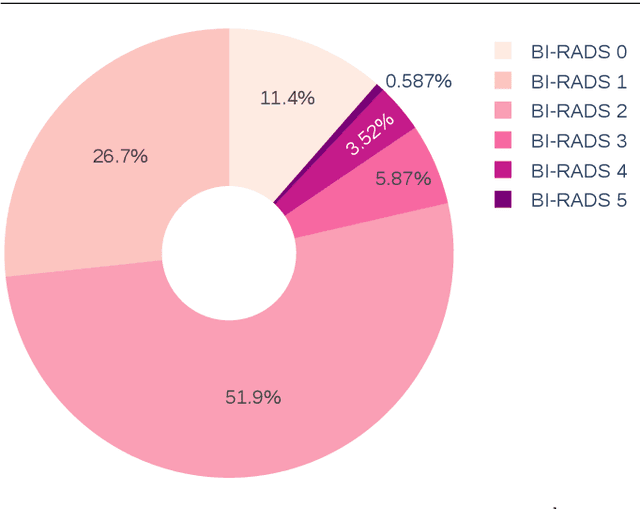
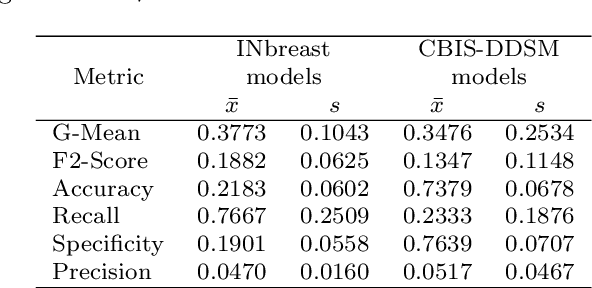
Abstract:The implementation of deep learning based computer aided diagnosis systems for the classification of mammogram images can help in improving the accuracy, reliability, and cost of diagnosing patients. However, training a deep learning model requires a considerable amount of labeled images, which can be expensive to obtain as time and effort from clinical practitioners is required. A number of publicly available datasets have been built with data from different hospitals and clinics. However, using models trained on these datasets for later work on images sampled from a different hospital or clinic might result in lower performance. This is due to the distribution mismatch of the datasets, which include different patient populations and image acquisition protocols. The scarcity of labeled data can also bring a challenge towards the application of transfer learning with models trained using these source datasets. In this work, a real world scenario is evaluated where a novel target dataset sampled from a private Costa Rican clinic is used, with few labels and heavily imbalanced data. The use of two popular and publicly available datasets (INbreast and CBIS-DDSM) as source data, to train and test the models on the novel target dataset, is evaluated. The use of the semi-supervised deep learning approach known as MixMatch, to leverage the usage of unlabeled data from the target dataset, is proposed and evaluated. In the tests, the performance of models is extensively measured, using different metrics to assess the performance of a classifier under heavy data imbalance conditions. It is shown that the use of semi-supervised deep learning combined with fine-tuning can provide a meaningful advantage when using scarce labeled observations. We make available the novel dataset for the benefit of the community.
Enforcing Morphological Information in Fully Convolutional Networks to Improve Cell Instance Segmentation in Fluorescence Microscopy Images
Jun 10, 2021
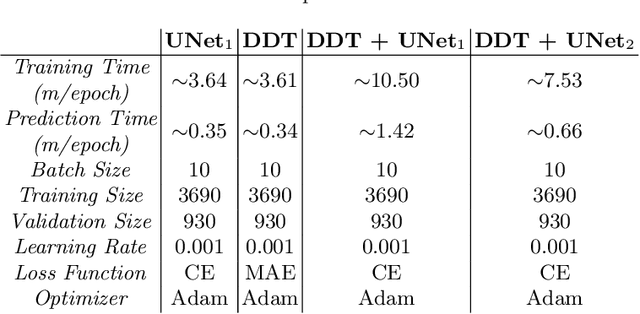
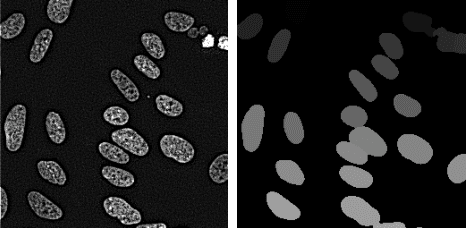
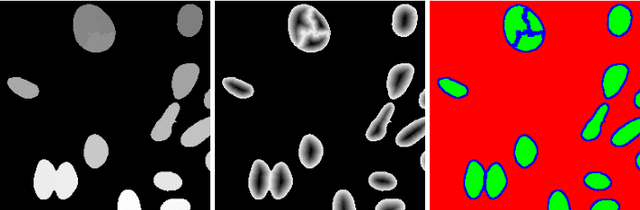
Abstract:Cell instance segmentation in fluorescence microscopy images is becoming essential for cancer dynamics and prognosis. Data extracted from cancer dynamics allows to understand and accurately model different metabolic processes such as proliferation. This enables customized and more precise cancer treatments. However, accurate cell instance segmentation, necessary for further cell tracking and behavior analysis, is still challenging in scenarios with high cell concentration and overlapping edges. Within this framework, we propose a novel cell instance segmentation approach based on the well-known U-Net architecture. To enforce the learning of morphological information per pixel, a deep distance transformer (DDT) acts as a back-bone model. The DDT output is subsequently used to train a top-model. The following top-models are considered: a three-class (\emph{e.g.,} foreground, background and cell border) U-net, and a watershed transform. The obtained results suggest a performance boost over traditional U-Net architectures. This opens an interesting research line around the idea of injecting morphological information into a fully convolutional model.
Correcting Data Imbalance for Semi-Supervised Covid-19 Detection Using X-ray Chest Images
Aug 20, 2020
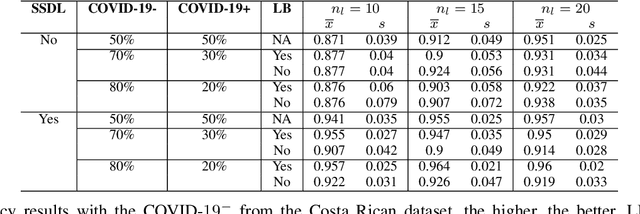
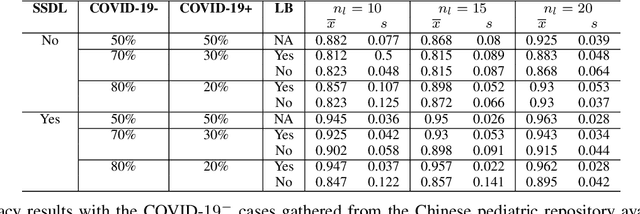
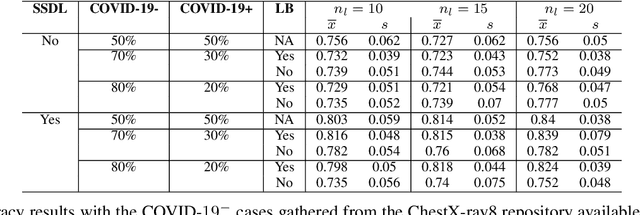
Abstract:The Corona Virus (COVID-19) is an internationalpandemic that has quickly propagated throughout the world. The application of deep learning for image classification of chest X-ray images of Covid-19 patients, could become a novel pre-diagnostic detection methodology. However, deep learning architectures require large labelled datasets. This is often a limitation when the subject of research is relatively new as in the case of the virus outbreak, where dealing with small labelled datasets is a challenge. Moreover, in the context of a new highly infectious disease, the datasets are also highly imbalanced,with few observations from positive cases of the new disease. In this work we evaluate the performance of the semi-supervised deep learning architecture known as MixMatch using a very limited number of labelled observations and highly imbalanced labelled dataset. We propose a simple approach for correcting data imbalance, re-weight each observationin the loss function, giving a higher weight to the observationscorresponding to the under-represented class. For unlabelled observations, we propose the usage of the pseudo and augmentedlabels calculated by MixMatch to choose the appropriate weight. The MixMatch method combined with the proposed pseudo-label based balance correction improved classification accuracy by up to 10%, with respect to the non balanced MixMatch algorithm, with statistical significance. We tested our proposed approach with several available datasets using 10, 15 and 20 labelledobservations. Additionally, a new dataset is included among thetested datasets, composed of chest X-ray images of Costa Rican adult patients
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge