Clément Vignac
Equivariant 3D-Conditional Diffusion Models for Molecular Linker Design
Oct 11, 2022



Abstract:Fragment-based drug discovery has been an effective paradigm in early-stage drug development. An open challenge in this area is designing linkers between disconnected molecular fragments of interest to obtain chemically-relevant candidate drug molecules. In this work, we propose DiffLinker, an E(3)-equivariant 3D-conditional diffusion model for molecular linker design. Given a set of disconnected fragments, our model places missing atoms in between and designs a molecule incorporating all the initial fragments. Unlike previous approaches that are only able to connect pairs of molecular fragments, our method can link an arbitrary number of fragments. Additionally, the model automatically determines the number of atoms in the linker and its attachment points to the input fragments. We demonstrate that DiffLinker outperforms other methods on the standard datasets generating more diverse and synthetically-accessible molecules. Besides, we experimentally test our method in real-world applications, showing that it can successfully generate valid linkers conditioned on target protein pockets.
Equivariant Diffusion for Molecule Generation in 3D
Mar 31, 2022



Abstract:This work introduces a diffusion model for molecule generation in 3D that is equivariant to Euclidean transformations. Our E(3) Equivariant Diffusion Model (EDM) learns to denoise a diffusion process with an equivariant network that jointly operates on both continuous (atom coordinates) and categorical features (atom types). In addition, we provide a probabilistic analysis which admits likelihood computation of molecules using our model. Experimentally, the proposed method significantly outperforms previous 3D molecular generative methods regarding the quality of generated samples and efficiency at training time.
Modurec: Recommender Systems with Feature and Time Modulation
Oct 13, 2020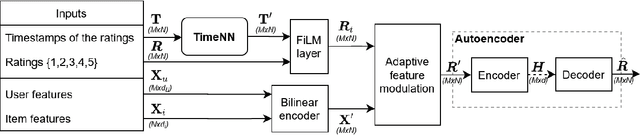
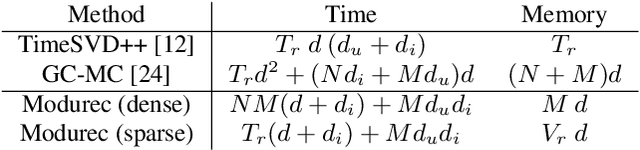

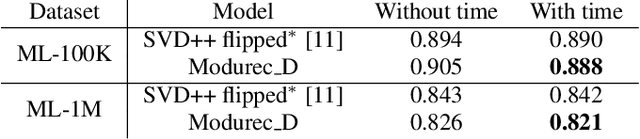
Abstract:Current state of the art algorithms for recommender systems are mainly based on collaborative filtering, which exploits user ratings to discover latent factors in the data. These algorithms unfortunately do not make effective use of other features, which can help solve two well identified problems of collaborative filtering: cold start (not enough data is available for new users or products) and concept shift (the distribution of ratings changes over time). To address these problems, we propose Modurec: an autoencoder-based method that combines all available information using the feature-wise modulation mechanism, which has demonstrated its effectiveness in several fields. While time information helps mitigate the effects of concept shift, the combination of user and item features improve prediction performance when little data is available. We show on Movielens datasets that these modifications produce state-of-the-art results in most evaluated settings compared with standard autoencoder-based methods and other collaborative filtering approaches.
On the choice of graph neural network architectures
Nov 13, 2019


Abstract:Seminal works on graph neural networks have primarily targeted semi-supervised node classification problems with few observed labels and high-dimensional signals. With the development of graph networks, this setup has become a de facto benchmark for a significant body of research. Interestingly, several works have recently shown that graph neural networks do not perform much better than predefined low-pass filters followed by a linear classifier in these particular settings. However, when learning with little data in a high-dimensional space, it is not surprising that simple and heavily regularized learning methods are near-optimal. In this paper, we show empirically that in settings with fewer features and more training data, more complex graph networks significantly outperform simpler architectures, and propose a few insights towards to the proper choice of graph neural networks architectures. We finally outline the importance of using sufficiently diverse benchmarks (including lower dimensional signals as well) when designing and studying new types of graph neural networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge