Chenyang Qiu
CAFusion: Controllable Anatomical Synthesis of Perirectal Lymph Nodes via SDF-guided Diffusion
Mar 10, 2025Abstract:Lesion synthesis methods have made significant progress in generating large-scale synthetic datasets. However, existing approaches predominantly focus on texture synthesis and often fail to accurately model masks for anatomically complex lesions. Additionally, these methods typically lack precise control over the synthesis process. For example, perirectal lymph nodes, which range in diameter from 1 mm to 10 mm, exhibit irregular and intricate contours that are challenging for current techniques to replicate faithfully. To address these limitations, we introduce CAFusion, a novel approach for synthesizing perirectal lymph nodes. By leveraging Signed Distance Functions (SDF), CAFusion generates highly realistic 3D anatomical structures. Furthermore, it offers flexible control over both anatomical and textural features by decoupling the generation of morphological attributes (such as shape, size, and position) from textural characteristics, including signal intensity. Experimental results demonstrate that our synthetic data substantially improve segmentation performance, achieving a 6.45% increase in the Dice coefficient. In the visual Turing test, experienced radiologists found it challenging to distinguish between synthetic and real lesions, highlighting the high degree of realism and anatomical accuracy achieved by our approach. These findings validate the effectiveness of our method in generating high-quality synthetic lesions for advancing medical image processing applications.
Rapid and Accurate Diagnosis of Acute Aortic Syndrome using Non-contrast CT: A Large-scale, Retrospective, Multi-center and AI-based Study
Jun 25, 2024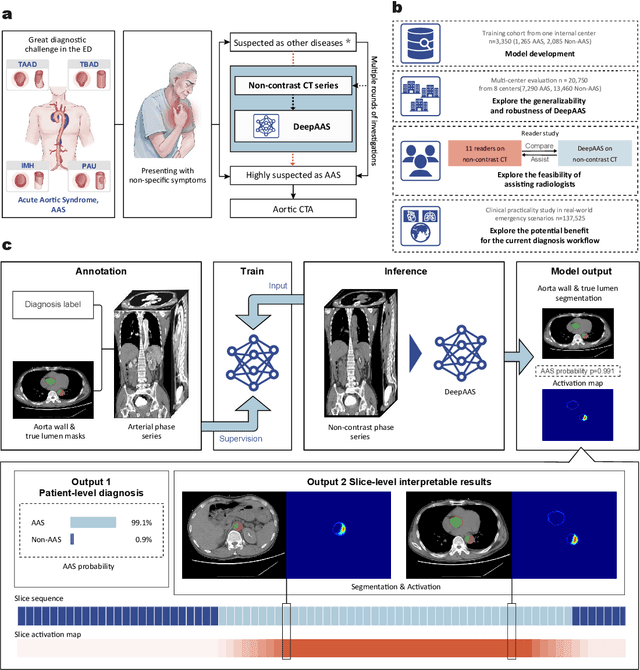
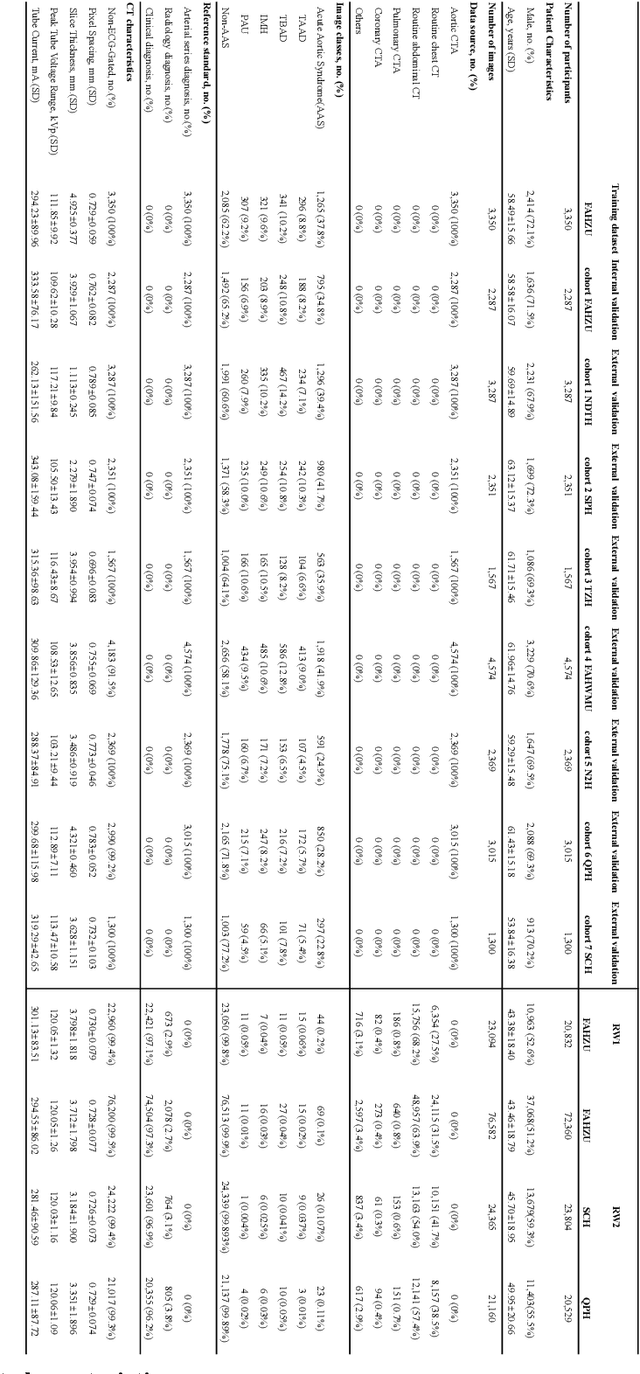
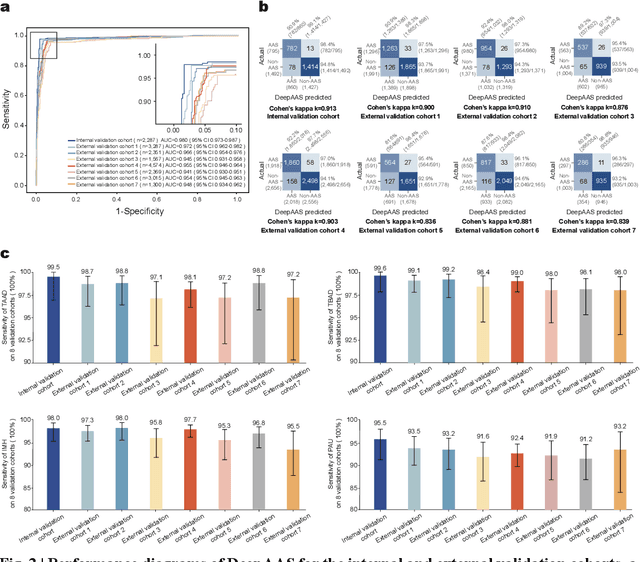
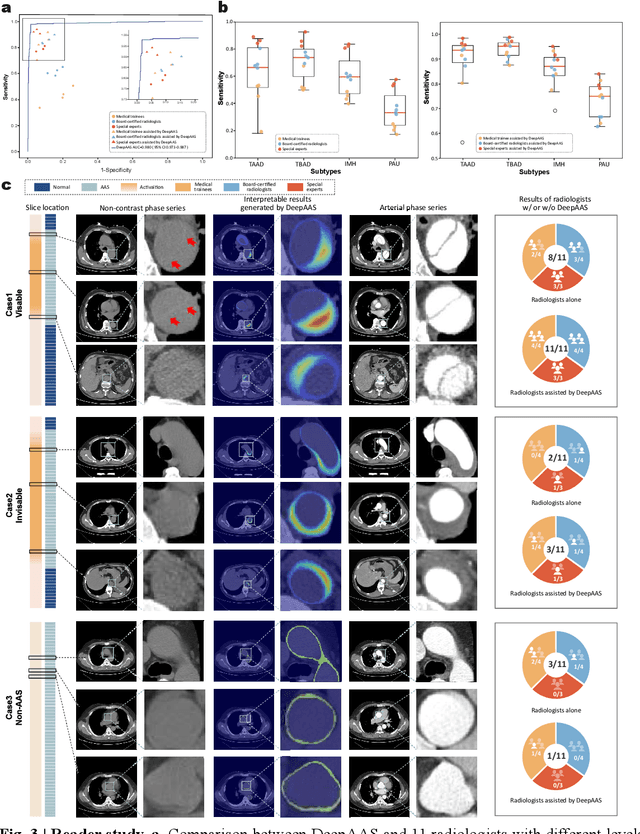
Abstract:Chest pain symptoms are highly prevalent in emergency departments (EDs), where acute aortic syndrome (AAS) is a catastrophic cardiovascular emergency with a high fatality rate, especially when timely and accurate treatment is not administered. However, current triage practices in the ED can cause up to approximately half of patients with AAS to have an initially missed diagnosis or be misdiagnosed as having other acute chest pain conditions. Subsequently, these AAS patients will undergo clinically inaccurate or suboptimal differential diagnosis. Fortunately, even under these suboptimal protocols, nearly all these patients underwent non-contrast CT covering the aorta anatomy at the early stage of differential diagnosis. In this study, we developed an artificial intelligence model (DeepAAS) using non-contrast CT, which is highly accurate for identifying AAS and provides interpretable results to assist in clinical decision-making. Performance was assessed in two major phases: a multi-center retrospective study (n = 20,750) and an exploration in real-world emergency scenarios (n = 137,525). In the multi-center cohort, DeepAAS achieved a mean area under the receiver operating characteristic curve of 0.958 (95% CI 0.950-0.967). In the real-world cohort, DeepAAS detected 109 AAS patients with misguided initial suspicion, achieving 92.6% (95% CI 76.2%-97.5%) in mean sensitivity and 99.2% (95% CI 99.1%-99.3%) in mean specificity. Our AI model performed well on non-contrast CT at all applicable early stages of differential diagnosis workflows, effectively reduced the overall missed diagnosis and misdiagnosis rate from 48.8% to 4.8% and shortened the diagnosis time for patients with misguided initial suspicion from an average of 681.8 (74-11,820) mins to 68.5 (23-195) mins. DeepAAS could effectively fill the gap in the current clinical workflow without requiring additional tests.
Meply: A Large-scale Dataset and Baseline Evaluations for Metastatic Perirectal Lymph Node Detection and Segmentation
Apr 13, 2024



Abstract:Accurate segmentation of metastatic lymph nodes in rectal cancer is crucial for the staging and treatment of rectal cancer. However, existing segmentation approaches face challenges due to the absence of pixel-level annotated datasets tailored for lymph nodes around the rectum. Additionally, metastatic lymph nodes are characterized by their relatively small size, irregular shapes, and lower contrast compared to the background, further complicating the segmentation task. To address these challenges, we present the first large-scale perirectal metastatic lymph node CT image dataset called Meply, which encompasses pixel-level annotations of 269 patients diagnosed with rectal cancer. Furthermore, we introduce a novel lymph-node segmentation model named CoSAM. The CoSAM utilizes sequence-based detection to guide the segmentation of metastatic lymph nodes in rectal cancer, contributing to improved localization performance for the segmentation model. It comprises three key components: sequence-based detection module, segmentation module, and collaborative convergence unit. To evaluate the effectiveness of CoSAM, we systematically compare its performance with several popular segmentation methods using the Meply dataset. Our code and dataset will be publicly available at: https://github.com/kanydao/CoSAM.
Refining Latent Homophilic Structures over Heterophilic Graphs for Robust Graph Convolution Networks
Dec 27, 2023



Abstract:Graph convolution networks (GCNs) are extensively utilized in various graph tasks to mine knowledge from spatial data. Our study marks the pioneering attempt to quantitatively investigate the GCN robustness over omnipresent heterophilic graphs for node classification. We uncover that the predominant vulnerability is caused by the structural out-of-distribution (OOD) issue. This finding motivates us to present a novel method that aims to harden GCNs by automatically learning Latent Homophilic Structures over heterophilic graphs. We term such a methodology as LHS. To elaborate, our initial step involves learning a latent structure by employing a novel self-expressive technique based on multi-node interactions. Subsequently, the structure is refined using a pairwisely constrained dual-view contrastive learning approach. We iteratively perform the above procedure, enabling a GCN model to aggregate information in a homophilic way on heterophilic graphs. Armed with such an adaptable structure, we can properly mitigate the structural OOD threats over heterophilic graphs. Experiments on various benchmarks show the effectiveness of the proposed LHS approach for robust GCNs.
CARE: A Large Scale CT Image Dataset and Clinical Applicable Benchmark Model for Rectal Cancer Segmentation
Aug 16, 2023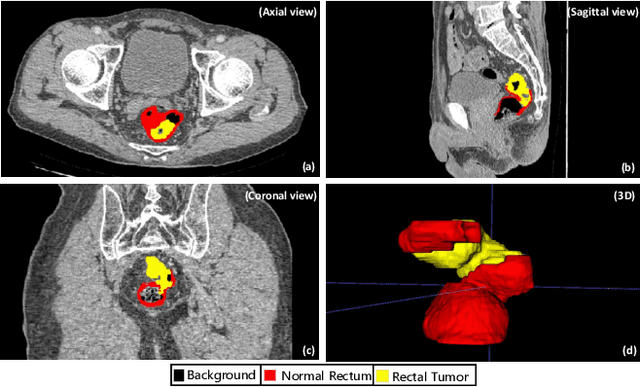



Abstract:Rectal cancer segmentation of CT image plays a crucial role in timely clinical diagnosis, radiotherapy treatment, and follow-up. Although current segmentation methods have shown promise in delineating cancerous tissues, they still encounter challenges in achieving high segmentation precision. These obstacles arise from the intricate anatomical structures of the rectum and the difficulties in performing differential diagnosis of rectal cancer. Additionally, a major obstacle is the lack of a large-scale, finely annotated CT image dataset for rectal cancer segmentation. To address these issues, this work introduces a novel large scale rectal cancer CT image dataset CARE with pixel-level annotations for both normal and cancerous rectum, which serves as a valuable resource for algorithm research and clinical application development. Moreover, we propose a novel medical cancer lesion segmentation benchmark model named U-SAM. The model is specifically designed to tackle the challenges posed by the intricate anatomical structures of abdominal organs by incorporating prompt information. U-SAM contains three key components: promptable information (e.g., points) to aid in target area localization, a convolution module for capturing low-level lesion details, and skip-connections to preserve and recover spatial information during the encoding-decoding process. To evaluate the effectiveness of U-SAM, we systematically compare its performance with several popular segmentation methods on the CARE dataset. The generalization of the model is further verified on the WORD dataset. Extensive experiments demonstrate that the proposed U-SAM outperforms state-of-the-art methods on these two datasets. These experiments can serve as the baseline for future research and clinical application development.
Fast Community Detection based on Graph Autoencoder Reconstruction
Mar 07, 2022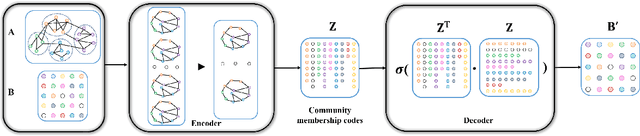
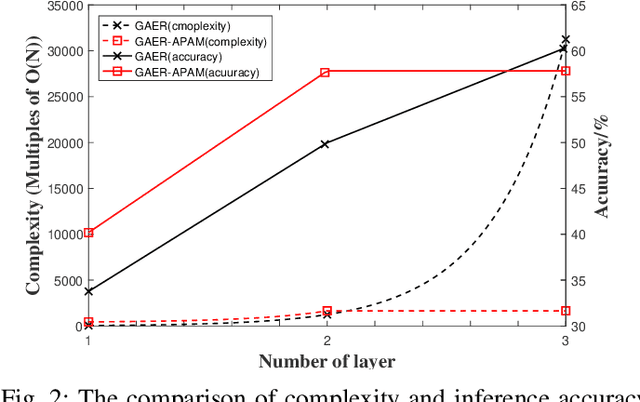
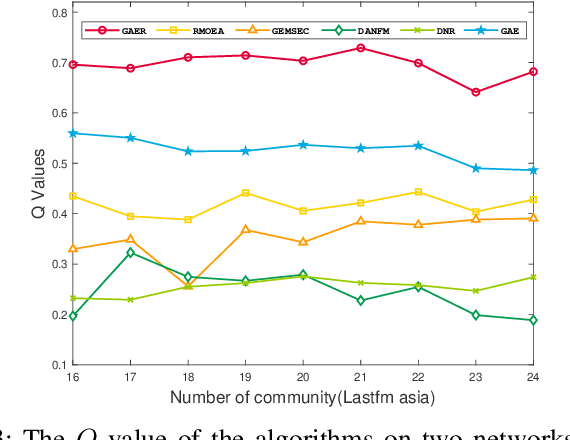
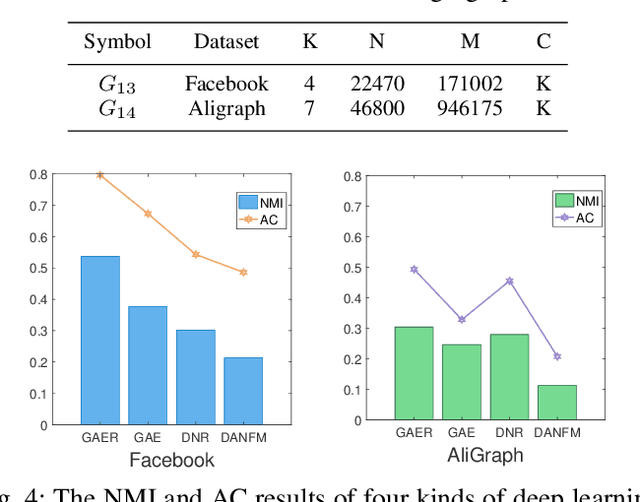
Abstract:With the rapid development of big data, how to efficiently and accurately discover tight community structures in large-scale networks for knowledge discovery has attracted more and more attention. In this paper, a community detection framework based on Graph AutoEncoder Reconstruction (noted as GAER) is proposed for the first time. GAER is a highly scalable framework which does not require any prior information. We decompose the graph autoencoder-based one-step encoding into the two-stage encoding framework to adapt to the real-world big data system by reducing complexity from the original O(N^2) to O(N). At the same time, based on the advantages of GAER support module plug-and-play configuration and incremental community detection, we further propose a peer awareness based module for real-time large graphs, which can realize the new nodes community detection at a faster speed, and accelerate model inference with the 6.15 times - 14.03 times speed. Finally, we apply the GAER on multiple real-world datasets, including some large-scale networks. The experimental result verified that GAER has achieved the superior performance on almost all networks.
VGAER: graph neural network reconstruction based community detection
Jan 29, 2022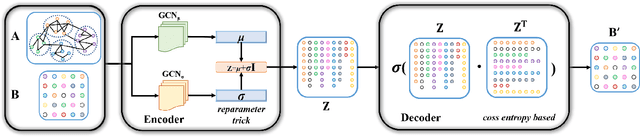
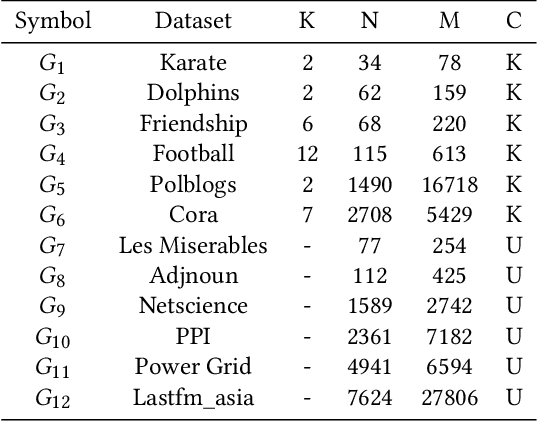
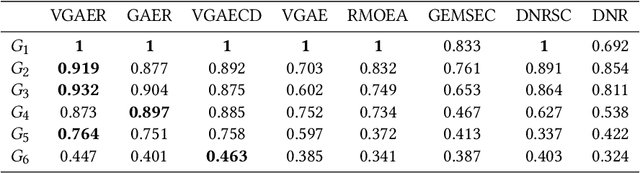
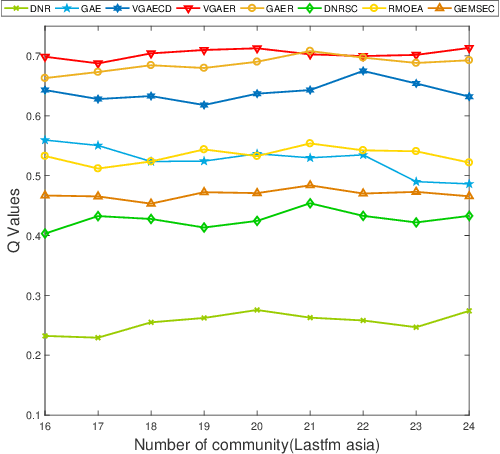
Abstract:Community detection is a fundamental and important issue in network science, but there are only a few community detection algorithms based on graph neural networks, among which unsupervised algorithms are almost blank. By fusing the high-order modularity information with network features, this paper proposes a Variational Graph AutoEncoder Reconstruction based community detection VGAER for the first time, and gives its non-probabilistic version. They do not need any prior information. We have carefully designed corresponding input features, decoder, and downstream tasks based on the community detection task and these designs are concise, natural, and perform well (NMI values under our design are improved by 59.1% - 565.9%). Based on a series of experiments with wide range of datasets and advanced methods, VGAER has achieved superior performance and shows strong competitiveness and potential with a simpler design. Finally, we report the results of algorithm convergence analysis and t-SNE visualization, which clearly depicted the stable performance and powerful network modularity ability of VGAER. Our codes are available at https://github.com/qcydm/VGAER.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge