Wanqin Wang
CAFusion: Controllable Anatomical Synthesis of Perirectal Lymph Nodes via SDF-guided Diffusion
Mar 10, 2025Abstract:Lesion synthesis methods have made significant progress in generating large-scale synthetic datasets. However, existing approaches predominantly focus on texture synthesis and often fail to accurately model masks for anatomically complex lesions. Additionally, these methods typically lack precise control over the synthesis process. For example, perirectal lymph nodes, which range in diameter from 1 mm to 10 mm, exhibit irregular and intricate contours that are challenging for current techniques to replicate faithfully. To address these limitations, we introduce CAFusion, a novel approach for synthesizing perirectal lymph nodes. By leveraging Signed Distance Functions (SDF), CAFusion generates highly realistic 3D anatomical structures. Furthermore, it offers flexible control over both anatomical and textural features by decoupling the generation of morphological attributes (such as shape, size, and position) from textural characteristics, including signal intensity. Experimental results demonstrate that our synthetic data substantially improve segmentation performance, achieving a 6.45% increase in the Dice coefficient. In the visual Turing test, experienced radiologists found it challenging to distinguish between synthetic and real lesions, highlighting the high degree of realism and anatomical accuracy achieved by our approach. These findings validate the effectiveness of our method in generating high-quality synthetic lesions for advancing medical image processing applications.
LN-Gen: Rectal Lymph Nodes Generation via Anatomical Features
Aug 27, 2024



Abstract:Accurate segmentation of rectal lymph nodes is crucial for the staging and treatment planning of rectal cancer. However, the complexity of the surrounding anatomical structures and the scarcity of annotated data pose significant challenges. This study introduces a novel lymph node synthesis technique aimed at generating diverse and realistic synthetic rectal lymph node samples to mitigate the reliance on manual annotation. Unlike direct diffusion methods, which often produce masks that are discontinuous and of suboptimal quality, our approach leverages an implicit SDF-based method for mask generation, ensuring the production of continuous, stable, and morphologically diverse masks. Experimental results demonstrate that our synthetic data significantly improves segmentation performance. Our work highlights the potential of diffusion model for accurately synthesizing structurally complex lesions, such as lymph nodes in rectal cancer, alleviating the challenge of limited annotated data in this field and aiding in advancements in rectal cancer diagnosis and treatment.
Meply: A Large-scale Dataset and Baseline Evaluations for Metastatic Perirectal Lymph Node Detection and Segmentation
Apr 13, 2024



Abstract:Accurate segmentation of metastatic lymph nodes in rectal cancer is crucial for the staging and treatment of rectal cancer. However, existing segmentation approaches face challenges due to the absence of pixel-level annotated datasets tailored for lymph nodes around the rectum. Additionally, metastatic lymph nodes are characterized by their relatively small size, irregular shapes, and lower contrast compared to the background, further complicating the segmentation task. To address these challenges, we present the first large-scale perirectal metastatic lymph node CT image dataset called Meply, which encompasses pixel-level annotations of 269 patients diagnosed with rectal cancer. Furthermore, we introduce a novel lymph-node segmentation model named CoSAM. The CoSAM utilizes sequence-based detection to guide the segmentation of metastatic lymph nodes in rectal cancer, contributing to improved localization performance for the segmentation model. It comprises three key components: sequence-based detection module, segmentation module, and collaborative convergence unit. To evaluate the effectiveness of CoSAM, we systematically compare its performance with several popular segmentation methods using the Meply dataset. Our code and dataset will be publicly available at: https://github.com/kanydao/CoSAM.
CARE: A Large Scale CT Image Dataset and Clinical Applicable Benchmark Model for Rectal Cancer Segmentation
Aug 16, 2023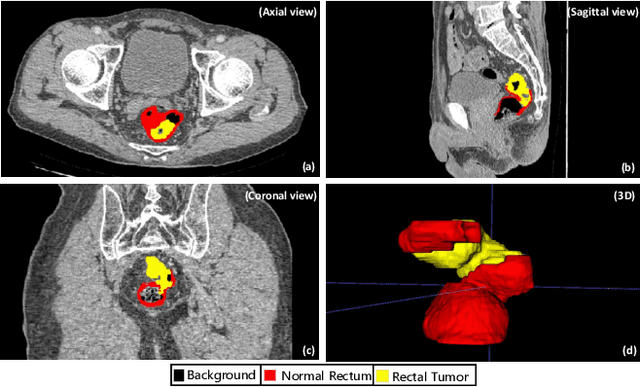



Abstract:Rectal cancer segmentation of CT image plays a crucial role in timely clinical diagnosis, radiotherapy treatment, and follow-up. Although current segmentation methods have shown promise in delineating cancerous tissues, they still encounter challenges in achieving high segmentation precision. These obstacles arise from the intricate anatomical structures of the rectum and the difficulties in performing differential diagnosis of rectal cancer. Additionally, a major obstacle is the lack of a large-scale, finely annotated CT image dataset for rectal cancer segmentation. To address these issues, this work introduces a novel large scale rectal cancer CT image dataset CARE with pixel-level annotations for both normal and cancerous rectum, which serves as a valuable resource for algorithm research and clinical application development. Moreover, we propose a novel medical cancer lesion segmentation benchmark model named U-SAM. The model is specifically designed to tackle the challenges posed by the intricate anatomical structures of abdominal organs by incorporating prompt information. U-SAM contains three key components: promptable information (e.g., points) to aid in target area localization, a convolution module for capturing low-level lesion details, and skip-connections to preserve and recover spatial information during the encoding-decoding process. To evaluate the effectiveness of U-SAM, we systematically compare its performance with several popular segmentation methods on the CARE dataset. The generalization of the model is further verified on the WORD dataset. Extensive experiments demonstrate that the proposed U-SAM outperforms state-of-the-art methods on these two datasets. These experiments can serve as the baseline for future research and clinical application development.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge