CARE: A Large Scale CT Image Dataset and Clinical Applicable Benchmark Model for Rectal Cancer Segmentation
Paper and Code
Aug 16, 2023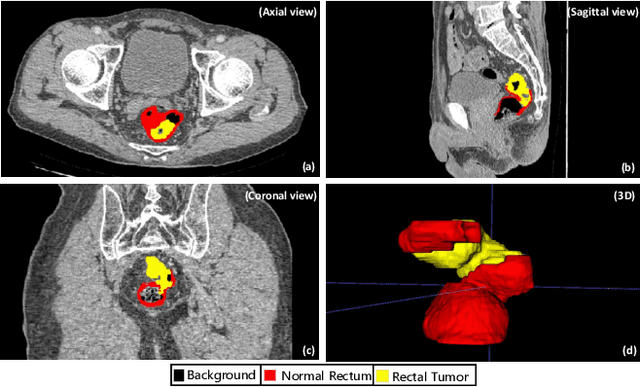



Rectal cancer segmentation of CT image plays a crucial role in timely clinical diagnosis, radiotherapy treatment, and follow-up. Although current segmentation methods have shown promise in delineating cancerous tissues, they still encounter challenges in achieving high segmentation precision. These obstacles arise from the intricate anatomical structures of the rectum and the difficulties in performing differential diagnosis of rectal cancer. Additionally, a major obstacle is the lack of a large-scale, finely annotated CT image dataset for rectal cancer segmentation. To address these issues, this work introduces a novel large scale rectal cancer CT image dataset CARE with pixel-level annotations for both normal and cancerous rectum, which serves as a valuable resource for algorithm research and clinical application development. Moreover, we propose a novel medical cancer lesion segmentation benchmark model named U-SAM. The model is specifically designed to tackle the challenges posed by the intricate anatomical structures of abdominal organs by incorporating prompt information. U-SAM contains three key components: promptable information (e.g., points) to aid in target area localization, a convolution module for capturing low-level lesion details, and skip-connections to preserve and recover spatial information during the encoding-decoding process. To evaluate the effectiveness of U-SAM, we systematically compare its performance with several popular segmentation methods on the CARE dataset. The generalization of the model is further verified on the WORD dataset. Extensive experiments demonstrate that the proposed U-SAM outperforms state-of-the-art methods on these two datasets. These experiments can serve as the baseline for future research and clinical application development.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge