Chengwei Ye
Is Visual Realism Enough? Evaluating Gait Biometric Fidelity in Generative AI Human Animation
Dec 22, 2025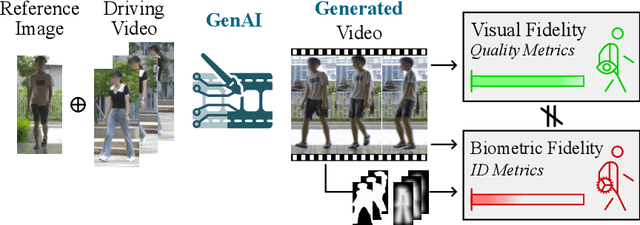
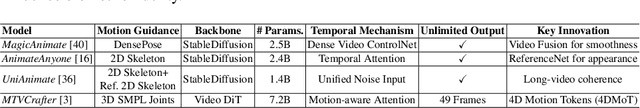
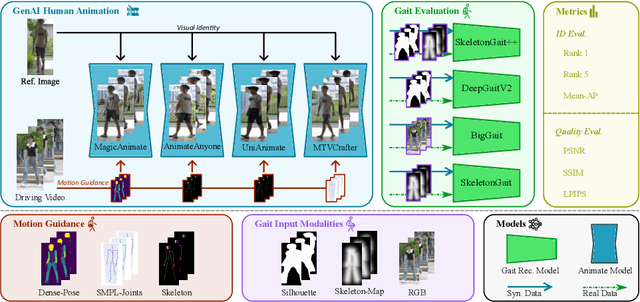
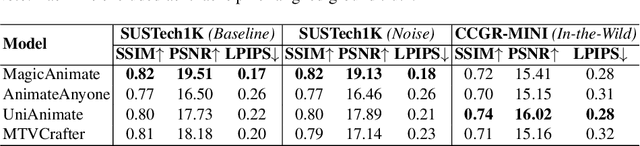
Abstract:Generative AI (GenAI) models have revolutionized animation, enabling the synthesis of humans and motion patterns with remarkable visual fidelity. However, generating truly realistic human animation remains a formidable challenge, where even minor inconsistencies can make a subject appear unnatural. This limitation is particularly critical when AI-generated videos are evaluated for behavioral biometrics, where subtle motion cues that define identity are easily lost or distorted. The present study investigates whether state-of-the-art GenAI human animation models can preserve the subtle spatio-temporal details needed for person identification through gait biometrics. Specifically, we evaluate four different GenAI models across two primary evaluation tasks to assess their ability to i) restore gait patterns from reference videos under varying conditions of complexity, and ii) transfer these gait patterns to different visual identities. Our results show that while visual quality is mostly high, biometric fidelity remains low in tasks focusing on identification, suggesting that current GenAI models struggle to disentangle identity from motion. Furthermore, through an identity transfer task, we expose a fundamental flaw in appearance-based gait recognition: when texture is disentangled from motion, identification collapses, proving current GenAI models rely on visual attributes rather than temporal dynamics.
Non-contact Vital Signs Detection in Dynamic Environments
May 13, 2025


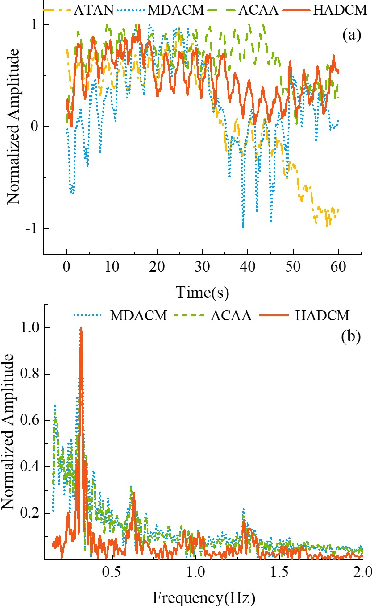
Abstract:Accurate phase demodulation is critical for vital sign detection using millimeter-wave radar. However, in complex environments, time-varying DC offsets and phase imbalances can severely degrade demodulation performance. To address this, we propose a novel DC offset calibration method alongside a Hilbert and Differential Cross-Multiply (HADCM) demodulation algorithm. The approach estimates time-varying DC offsets from neighboring signal peaks and valleys, then employs both differential forms and Hilbert transforms of the I/Q channel signals to extract vital sign information. Simulation and experimental results demonstrate that the proposed method maintains robust performance under low signal-to-noise ratios. Compared to existing demodulation techniques, it offers more accurate signal recovery in challenging scenarios and effectively suppresses noise interference.
CCL: Collaborative Curriculum Learning for Sparse-Reward Multi-Agent Reinforcement Learning via Co-evolutionary Task Evolution
May 08, 2025



Abstract:Sparse reward environments pose significant challenges in reinforcement learning, especially within multi-agent systems (MAS) where feedback is delayed and shared across agents, leading to suboptimal learning. We propose Collaborative Multi-dimensional Course Learning (CCL), a novel curriculum learning framework that addresses this by (1) refining intermediate tasks for individual agents, (2) using a variational evolutionary algorithm to generate informative subtasks, and (3) co-evolving agents with their environment to enhance training stability. Experiments on five cooperative tasks in the MPE and Hide-and-Seek environments show that CCL outperforms existing methods in sparse reward settings.
GaMNet: A Hybrid Network with Gabor Fusion and NMamba for Efficient 3D Glioma Segmentation
May 08, 2025Abstract:Gliomas are aggressive brain tumors that pose serious health risks. Deep learning aids in lesion segmentation, but CNN and Transformer-based models often lack context modeling or demand heavy computation, limiting real-time use on mobile medical devices. We propose GaMNet, integrating the NMamba module for global modeling and a multi-scale CNN for efficient local feature extraction. To improve interpretability and mimic the human visual system, we apply Gabor filters at multiple scales. Our method achieves high segmentation accuracy with fewer parameters and faster computation. Extensive experiments show GaMNet outperforms existing methods, notably reducing false positives and negatives, which enhances the reliability of clinical diagnosis.
GAME: Learning Multimodal Interactions via Graph Structures for Personality Trait Estimation
May 05, 2025Abstract:Apparent personality analysis from short videos poses significant chal-lenges due to the complex interplay of visual, auditory, and textual cues. In this paper, we propose GAME, a Graph-Augmented Multimodal Encoder designed to robustly model and fuse multi-source features for automatic personality prediction. For the visual stream, we construct a facial graph and introduce a dual-branch Geo Two-Stream Network, which combines Graph Convolutional Networks (GCNs) and Convolutional Neural Net-works (CNNs) with attention mechanisms to capture both structural and appearance-based facial cues. Complementing this, global context and iden-tity features are extracted using pretrained ResNet18 and VGGFace back-bones. To capture temporal dynamics, frame-level features are processed by a BiGRU enhanced with temporal attention modules. Meanwhile, audio representations are derived from the VGGish network, and linguistic se-mantics are captured via the XLM-Roberta transformer. To achieve effective multimodal integration, we propose a Channel Attention-based Fusion module, followed by a Multi-Layer Perceptron (MLP) regression head for predicting personality traits. Extensive experiments show that GAME con-sistently outperforms existing methods across multiple benchmarks, vali-dating its effectiveness and generalizability.
Graph-Driven Multimodal Feature Learning Framework for Apparent Personality Assessment
Apr 15, 2025Abstract:Predicting personality traits automatically has become a challenging problem in computer vision. This paper introduces an innovative multimodal feature learning framework for personality analysis in short video clips. For visual processing, we construct a facial graph and design a Geo-based two-stream network incorporating an attention mechanism, leveraging both Graph Convolutional Networks (GCN) and Convolutional Neural Networks (CNN) to capture static facial expressions. Additionally, ResNet18 and VGGFace networks are employed to extract global scene and facial appearance features at the frame level. To capture dynamic temporal information, we integrate a BiGRU with a temporal attention module for extracting salient frame representations. To enhance the model's robustness, we incorporate the VGGish CNN for audio-based features and XLM-Roberta for text-based features. Finally, a multimodal channel attention mechanism is introduced to integrate different modalities, and a Multi-Layer Perceptron (MLP) regression model is used to predict personality traits. Experimental results confirm that our proposed framework surpasses existing state-of-the-art approaches in performance.
Style Transfer Enabled Sim2Real Framework for Efficient Learning of Robotic Ultrasound Image Analysis Using Simulated Data
May 16, 2023Abstract:Robotic ultrasound (US) systems have shown great potential to make US examinations easier and more accurate. Recently, various machine learning techniques have been proposed to realize automatic US image interpretation for robotic US acquisition tasks. However, obtaining large amounts of real US imaging data for training is usually expensive or even unfeasible in some clinical applications. An alternative is to build a simulator to generate synthetic US data for training, but the differences between simulated and real US images may result in poor model performance. This work presents a Sim2Real framework to efficiently learn robotic US image analysis tasks based only on simulated data for real-world deployment. A style transfer module is proposed based on unsupervised contrastive learning and used as a preprocessing step to convert the real US images into the simulation style. Thereafter, a task-relevant model is designed to combine CNNs with vision transformers to generate the task-dependent prediction with improved generalization ability. We demonstrate the effectiveness of our method in an image regression task to predict the probe position based on US images in robotic transesophageal echocardiography (TEE). Our results show that using only simulated US data and a small amount of unlabelled real data for training, our method can achieve comparable performance to semi-supervised and fully supervised learning methods. Moreover, the effectiveness of our previously proposed CT-based US image simulation method is also indirectly confirmed.
Deep Learning-based Biological Anatomical Landmark Detection in Colonoscopy Videos
Aug 06, 2021



Abstract:Colonoscopy is a standard imaging tool for visualizing the entire gastrointestinal (GI) tract of patients to capture lesion areas. However, it takes the clinicians excessive time to review a large number of images extracted from colonoscopy videos. Thus, automatic detection of biological anatomical landmarks within the colon is highly demanded, which can help reduce the burden of clinicians by providing guidance information for the locations of lesion areas. In this article, we propose a novel deep learning-based approach to detect biological anatomical landmarks in colonoscopy videos. First, raw colonoscopy video sequences are pre-processed to reject interference frames. Second, a ResNet-101 based network is used to detect three biological anatomical landmarks separately to obtain the intermediate detection results. Third, to achieve more reliable localization of the landmark periods within the whole video period, we propose to post-process the intermediate detection results by identifying the incorrectly predicted frames based on their temporal distribution and reassigning them back to the correct class. Finally, the average detection accuracy reaches 99.75\%. Meanwhile, the average IoU of 0.91 shows a high degree of similarity between our predicted landmark periods and ground truth. The experimental results demonstrate that our proposed model is capable of accurately detecting and localizing biological anatomical landmarks from colonoscopy videos.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge