Charles L. Isbell
Perceptual Values from Observation
May 20, 2019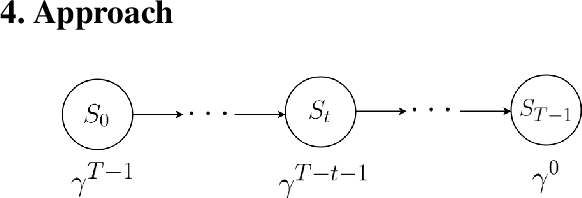
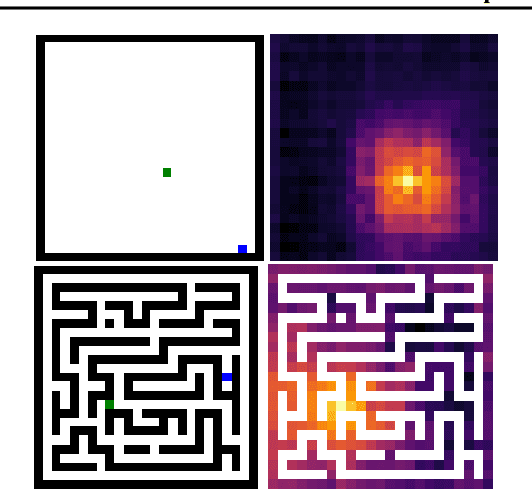
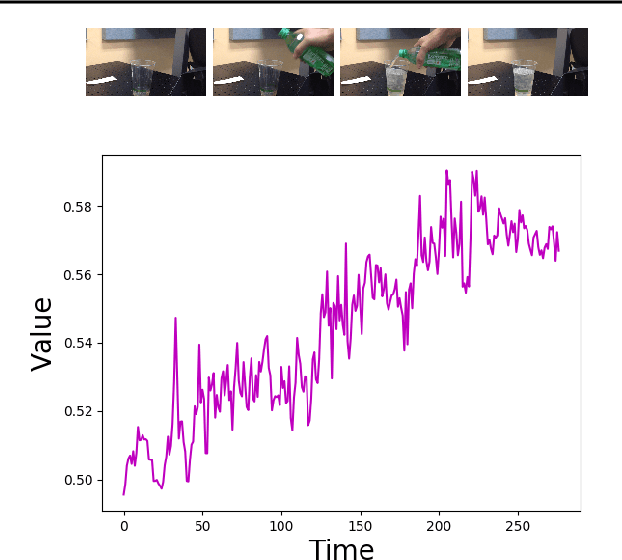
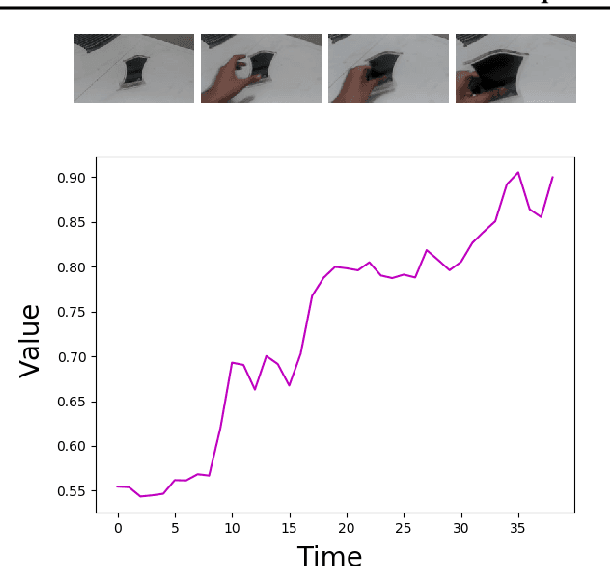
Abstract:Imitation by observation is an approach for learning from expert demonstrations that lack action information, such as videos. Recent approaches to this problem can be placed into two broad categories: training dynamics models that aim to predict the actions taken between states, and learning rewards or features for computing them for Reinforcement Learning (RL). In this paper, we introduce a novel approach that learns values, rather than rewards, directly from observations. We show that by using values, we can significantly speed up RL by removing the need to bootstrap action-values, as compared to sparse-reward specifications.
Purely Geometric Scene Association and Retrieval - A Case for Macro Scale 3D Geometry
Aug 03, 2018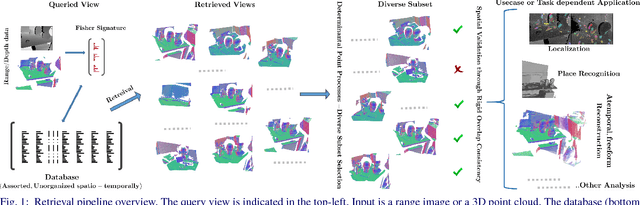
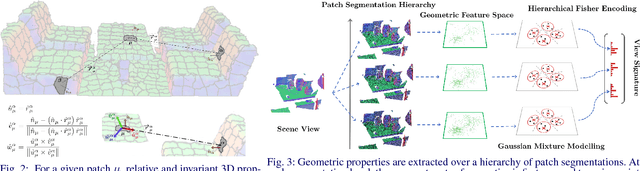
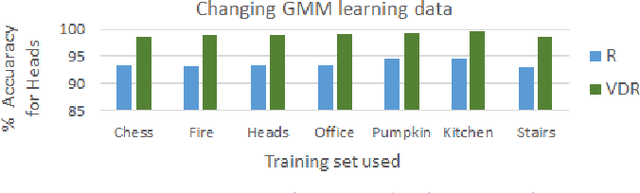
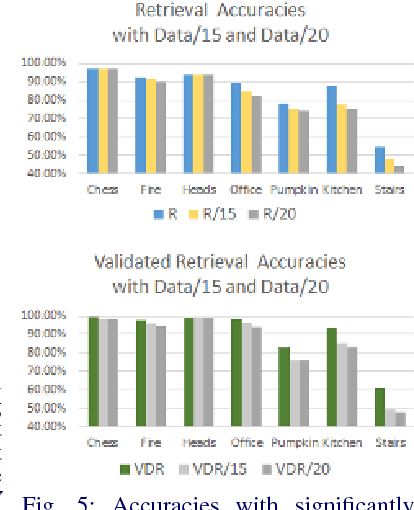
Abstract:We address the problems of measuring geometric similarity between 3D scenes, represented through point clouds or range data frames, and associating them. Our approach leverages macro-scale 3D structural geometry - the relative configuration of arbitrary surfaces and relationships among structures that are potentially far apart. We express such discriminative information in a viewpoint-invariant feature space. These are subsequently encoded in a frame-level signature that can be utilized to measure geometric similarity. Such a characterization is robust to noise, incomplete and partially overlapping data besides viewpoint changes. We show how it can be employed to select a diverse set of data frames which have structurally similar content, and how to validate whether views with similar geometric content are from the same scene. The problem is formulated as one of general purpose retrieval from an unannotated, spatio-temporally unordered database. Empirical analysis indicates that the presented approach thoroughly outperforms baselines on depth / range data. Its depth-only performance is competitive with state-of-the-art approaches with RGB or RGB-D inputs, including ones based on deep learning. Experiments show retrieval performance to hold up well with much sparser databases, which is indicative of the approach's robustness. The approach generalized well - it did not require dataset specific training, and scaled up in our experiments. Finally, we also demonstrate how geometrically diverse selection of views can result in richer 3D reconstructions.
Imitating Latent Policies from Observation
May 24, 2018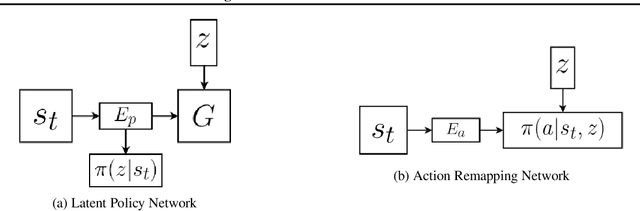
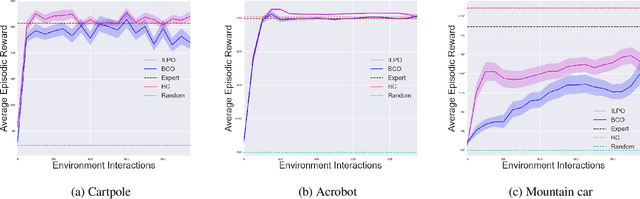
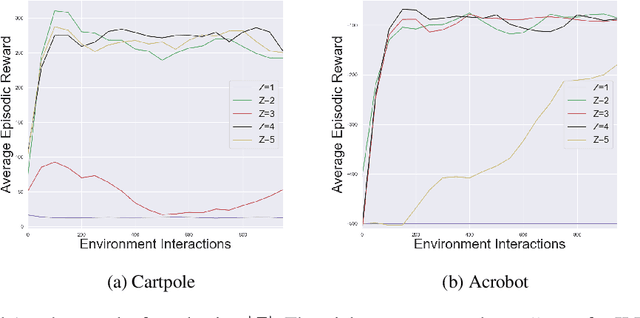
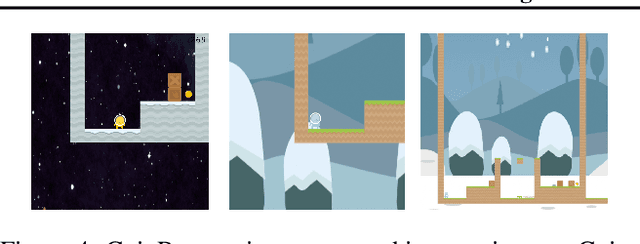
Abstract:We describe a novel approach to imitation learning that infers latent policies directly from state observations. We introduce a method that characterizes the causal effects of unknown actions on observations while simultaneously predicting their likelihood. We then outline an action alignment procedure that leverages a small amount of environment interactions to determine a mapping between latent and real-world actions. We show that this corrected labeling can be used for imitating the observed behavior, even though no expert actions are given. We evaluate our approach within classic control and photo-realistic visual environments and demonstrate that it performs well when compared to standard approaches.
MotifMark: Finding Regulatory Motifs in DNA Sequences
May 04, 2017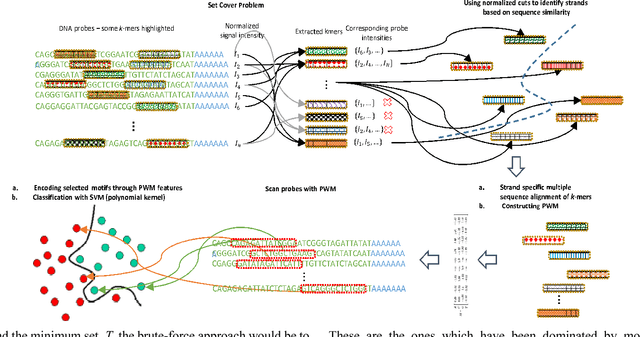
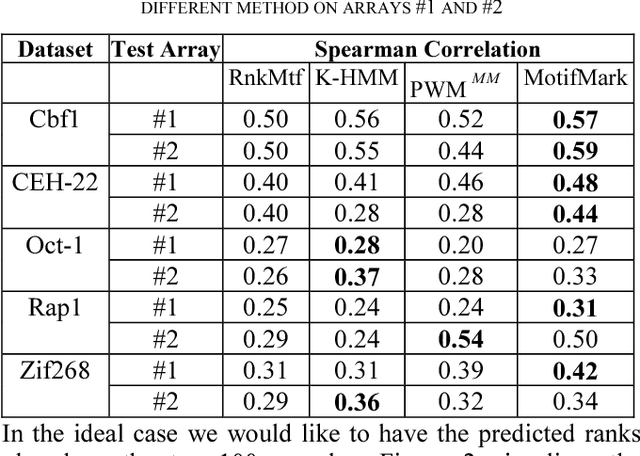
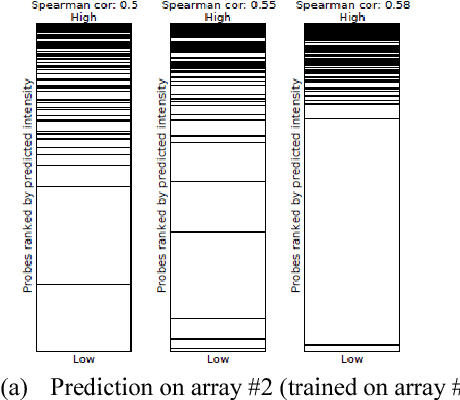
Abstract:The interaction between proteins and DNA is a key driving force in a significant number of biological processes such as transcriptional regulation, repair, recombination, splicing, and DNA modification. The identification of DNA-binding sites and the specificity of target proteins in binding to these regions are two important steps in understanding the mechanisms of these biological activities. A number of high-throughput technologies have recently emerged that try to quantify the affinity between proteins and DNA motifs. Despite their success, these technologies have their own limitations and fall short in precise characterization of motifs, and as a result, require further downstream analysis to extract useful and interpretable information from a haystack of noisy and inaccurate data. Here we propose MotifMark, a new algorithm based on graph theory and machine learning, that can find binding sites on candidate probes and rank their specificity in regard to the underlying transcription factor. We developed a pipeline to analyze experimental data derived from compact universal protein binding microarrays and benchmarked it against two of the most accurate motif search methods. Our results indicate that MotifMark can be a viable alternative technique for prediction of motif from protein binding microarrays and possibly other related high-throughput techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge