Chaonan Lin
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
COTR: Convolution in Transformer Network for End to End Polyp Detection
May 23, 2021
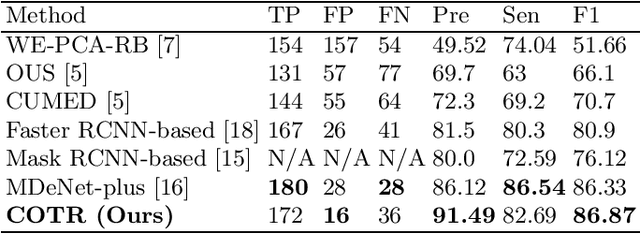
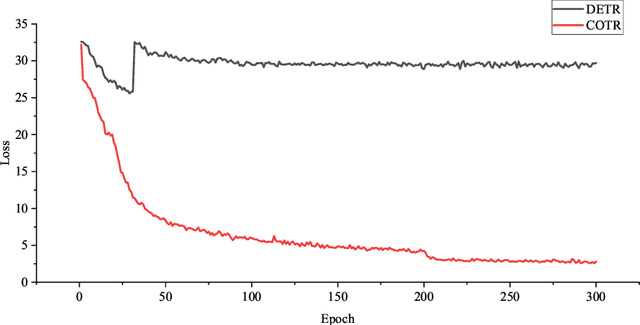
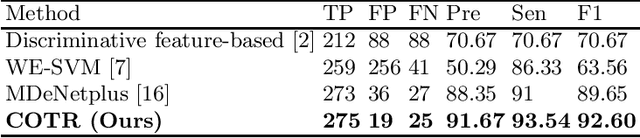
Abstract:Purpose: Colorectal cancer (CRC) is the second most common cause of cancer mortality worldwide. Colonoscopy is a widely used technique for colon screening and polyp lesions diagnosis. Nevertheless, manual screening using colonoscopy suffers from a substantial miss rate of polyps and is an overwhelming burden for endoscopists. Computer-aided diagnosis (CAD) for polyp detection has the potential to reduce human error and human burden. However, current polyp detection methods based on object detection framework need many handcrafted pre-processing and post-processing operations or user guidance that require domain-specific knowledge. Methods: In this paper, we propose a convolution in transformer (COTR) network for end-to-end polyp detection. Motivated by the detection transformer (DETR), COTR is constituted by a CNN for feature extraction, transformer encoder layers interleaved with convolutional layers for feature encoding and recalibration, transformer decoder layers for object querying, and a feed-forward network for detection prediction. Considering the slow convergence of DETR, COTR embeds convolution layers into transformer encoder for feature reconstruction and convergence acceleration. Results: Experimental results on two public polyp datasets show that COTR achieved 91.49\% precision, 82.69% sensitivity, and 86.87% F1-score on the ETIS-LARIB, and 91.67% precision, 93.54% sensitivity, and 92.60% F1-score on the CVC-ColonDB. Conclusion: This study proposed an end to end detection method based on detection transformer for colorectal polyp detection. Experimental results on ETIS-LARIB and CVC-ColonDB dataset demonstrated that the proposed model achieved comparable performance against state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge