Chao Che
Enhancing Autism Spectrum Disorder Early Detection with the Parent-Child Dyads Block-Play Protocol and an Attention-enhanced GCN-xLSTM Hybrid Deep Learning Framework
Aug 29, 2024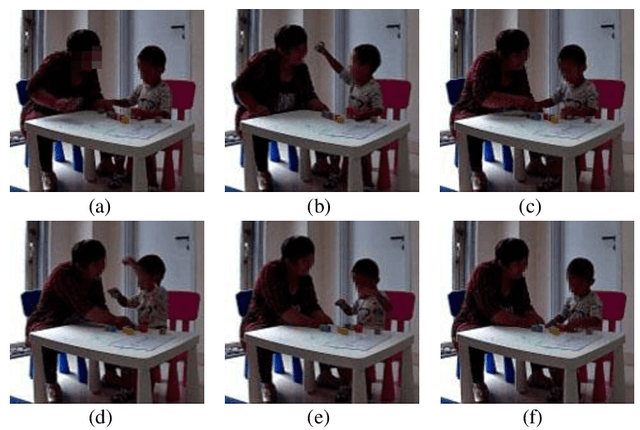
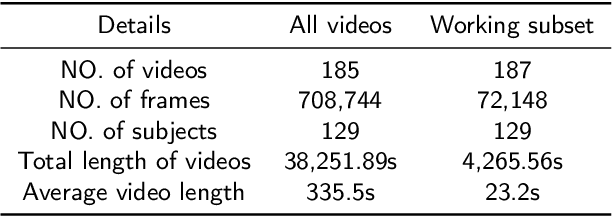

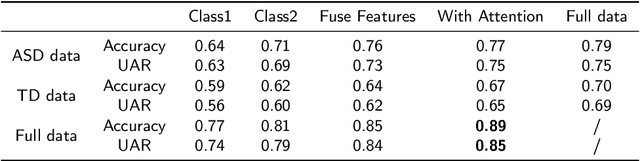
Abstract:Autism Spectrum Disorder (ASD) is a rapidly growing neurodevelopmental disorder. Performing a timely intervention is crucial for the growth of young children with ASD, but traditional clinical screening methods lack objectivity. This study introduces an innovative approach to early detection of ASD. The contributions are threefold. First, this work proposes a novel Parent-Child Dyads Block-Play (PCB) protocol, grounded in kinesiological and neuroscientific research, to identify behavioral patterns distinguishing ASD from typically developing (TD) toddlers. Second, we have compiled a substantial video dataset, featuring 40 ASD and 89 TD toddlers engaged in block play with parents. This dataset exceeds previous efforts on both the scale of participants and the length of individual sessions. Third, our approach to action analysis in videos employs a hybrid deep learning framework, integrating a two-stream graph convolution network with attention-enhanced xLSTM (2sGCN-AxLSTM). This framework is adept at capturing dynamic interactions between toddlers and parents by extracting spatial features correlated with upper body and head movements and focusing on global contextual information of action sequences over time. By learning these global features with spatio-temporal correlations, our 2sGCN-AxLSTM effectively analyzes dynamic human behavior patterns and demonstrates an unprecedented accuracy of 89.6\% in early detection of ASD. Our approach shows strong potential for enhancing early ASD diagnosis by accurately analyzing parent-child interactions, providing a critical tool to support timely and informed clinical decision-making.
JSCDS: A Core Data Selection Method with Jason-Shannon Divergence for Caries RGB Images-Efficient Learning
Jun 29, 2024



Abstract:Deep learning-based RGB caries detection improves the efficiency of caries identification and is crucial for preventing oral diseases. The performance of deep learning models depends on high-quality data and requires substantial training resources, making efficient deployment challenging. Core data selection, by eliminating low-quality and confusing data, aims to enhance training efficiency without significantly compromising model performance. However, distance-based data selection methods struggle to distinguish dependencies among high-dimensional caries data. To address this issue, we propose a Core Data Selection Method with Jensen-Shannon Divergence (JSCDS) for efficient caries image learning and caries classification. We describe the core data selection criterion as the distribution of samples in different classes. JSCDS calculates the cluster centers by sample embedding representation in the caries classification network and utilizes Jensen-Shannon Divergence to compute the mutual information between data samples and cluster centers, capturing nonlinear dependencies among high-dimensional data. The average mutual information is calculated to fit the above distribution, serving as the criterion for constructing the core set for model training. Extensive experiments on RGB caries datasets show that JSCDS outperforms other data selection methods in prediction performance and time consumption. Notably, JSCDS exceeds the performance of the full dataset model with only 50% of the core data, with its performance advantage becoming more pronounced in the 70% of core data.
The NIPS'17 Competition: A Multi-View Ensemble Classification Model for Clinically Actionable Genetic Mutations
Jun 26, 2018
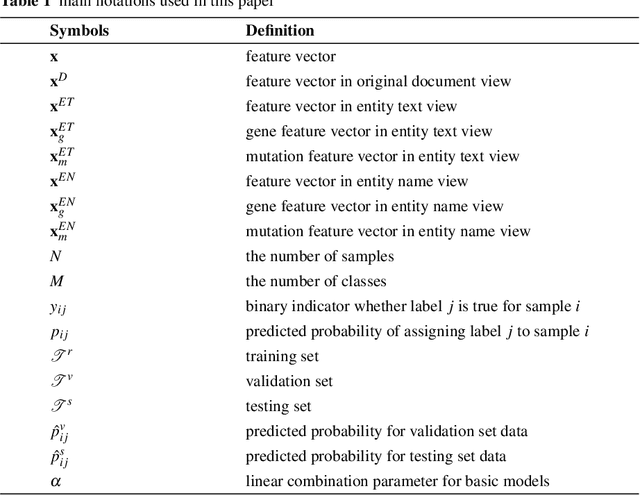
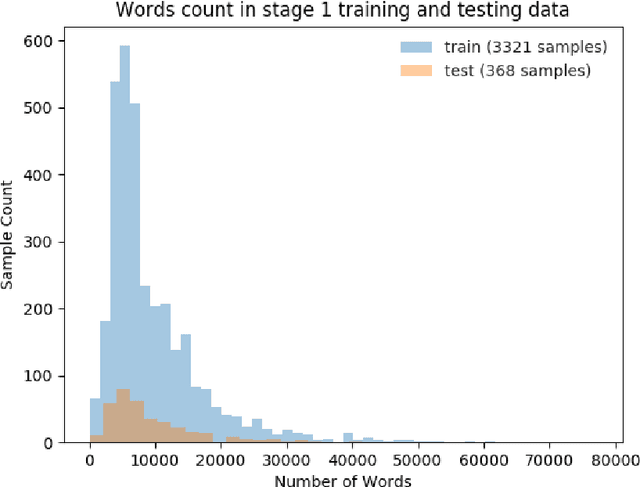
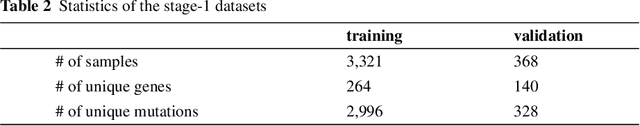
Abstract:This paper presents details of our winning solutions to the task IV of NIPS 2017 Competition Track entitled Classifying Clinically Actionable Genetic Mutations. The machine learning task aims to classify genetic mutations based on text evidence from clinical literature with promising performance. We develop a novel multi-view machine learning framework with ensemble classification models to solve the problem. During the Challenge, feature combinations derived from three views including document view, entity text view, and entity name view, which complements each other, are comprehensively explored. As the final solution, we submitted an ensemble of nine basic gradient boosting models which shows the best performance in the evaluation. The approach scores 0.5506 and 0.6694 in terms of logarithmic loss on a fixed split in stage-1 testing phase and 5-fold cross validation respectively, which also makes us ranked as a top-1 team out of more than 1,300 solutions in NIPS 2017 Competition Track IV.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge