Yongjun Zhu
The Effect of Gender Diversity on Scientific Team Impact: A Team Roles Perspective
Dec 29, 2025Abstract:The influence of gender diversity on the success of scientific teams is of great interest to academia. However, prior findings remain inconsistent, and most studies operationalize diversity in aggregate terms, overlooking internal role differentiation. This limitation obscures a more nuanced understanding of how gender diversity shapes team impact. In particular, the effect of gender diversity across different team roles remains poorly understood. To this end, we define a scientific team as all coauthors of a paper and measure team impact through five-year citation counts. Using author contribution statements, we classified members into leadership and support roles. Drawing on more than 130,000 papers from PLOS journals, most of which are in biomedical-related disciplines, we employed multivariable regression to examine the association between gender diversity in these roles and team impact. Furthermore, we apply a threshold regression model to investigate how team size moderates this relationship. The results show that (1) the relationship between gender diversity and team impact follows an inverted U-shape for both leadership and support groups; (2) teams with an all-female leadership group and an all-male support group achieve higher impact than other team types. Interestingly, (3) the effect of leadership-group gender diversity is significantly negative for small teams but becomes positive and statistically insignificant in large teams. In contrast, the estimates for support-group gender diversity remain significant and positive, regardless of team size.
JSCDS: A Core Data Selection Method with Jason-Shannon Divergence for Caries RGB Images-Efficient Learning
Jun 29, 2024



Abstract:Deep learning-based RGB caries detection improves the efficiency of caries identification and is crucial for preventing oral diseases. The performance of deep learning models depends on high-quality data and requires substantial training resources, making efficient deployment challenging. Core data selection, by eliminating low-quality and confusing data, aims to enhance training efficiency without significantly compromising model performance. However, distance-based data selection methods struggle to distinguish dependencies among high-dimensional caries data. To address this issue, we propose a Core Data Selection Method with Jensen-Shannon Divergence (JSCDS) for efficient caries image learning and caries classification. We describe the core data selection criterion as the distribution of samples in different classes. JSCDS calculates the cluster centers by sample embedding representation in the caries classification network and utilizes Jensen-Shannon Divergence to compute the mutual information between data samples and cluster centers, capturing nonlinear dependencies among high-dimensional data. The average mutual information is calculated to fit the above distribution, serving as the criterion for constructing the core set for model training. Extensive experiments on RGB caries datasets show that JSCDS outperforms other data selection methods in prediction performance and time consumption. Notably, JSCDS exceeds the performance of the full dataset model with only 50% of the core data, with its performance advantage becoming more pronounced in the 70% of core data.
The NIPS'17 Competition: A Multi-View Ensemble Classification Model for Clinically Actionable Genetic Mutations
Jun 26, 2018
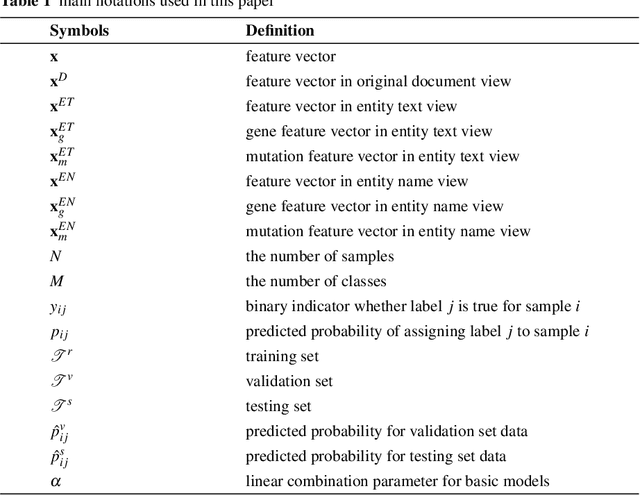
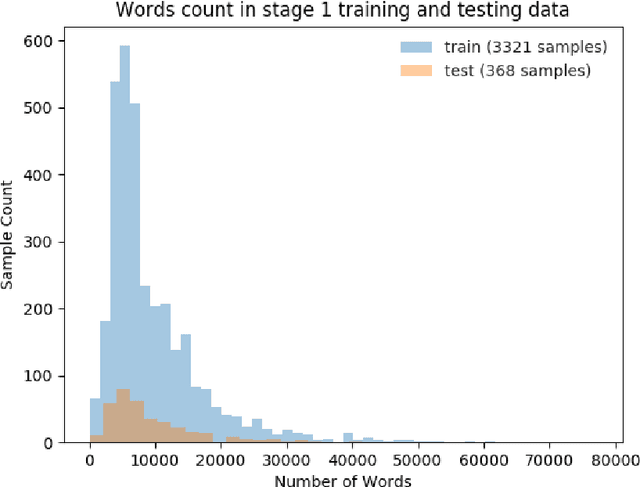
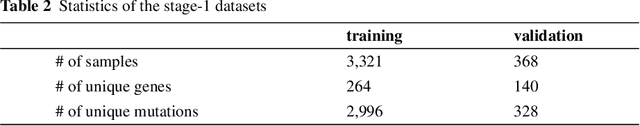
Abstract:This paper presents details of our winning solutions to the task IV of NIPS 2017 Competition Track entitled Classifying Clinically Actionable Genetic Mutations. The machine learning task aims to classify genetic mutations based on text evidence from clinical literature with promising performance. We develop a novel multi-view machine learning framework with ensemble classification models to solve the problem. During the Challenge, feature combinations derived from three views including document view, entity text view, and entity name view, which complements each other, are comprehensively explored. As the final solution, we submitted an ensemble of nine basic gradient boosting models which shows the best performance in the evaluation. The approach scores 0.5506 and 0.6694 in terms of logarithmic loss on a fixed split in stage-1 testing phase and 5-fold cross validation respectively, which also makes us ranked as a top-1 team out of more than 1,300 solutions in NIPS 2017 Competition Track IV.
A natural language interface to a graph-based bibliographic information retrieval system
Dec 10, 2016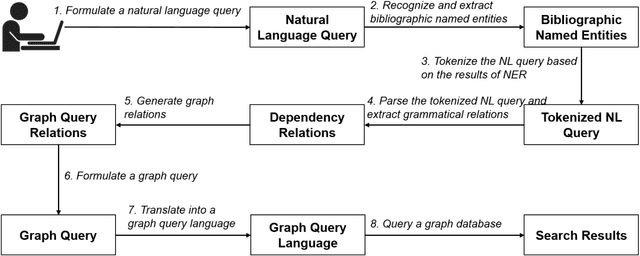

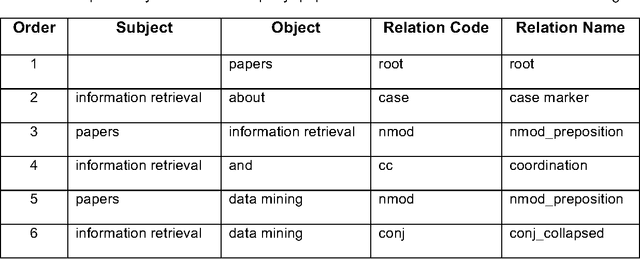
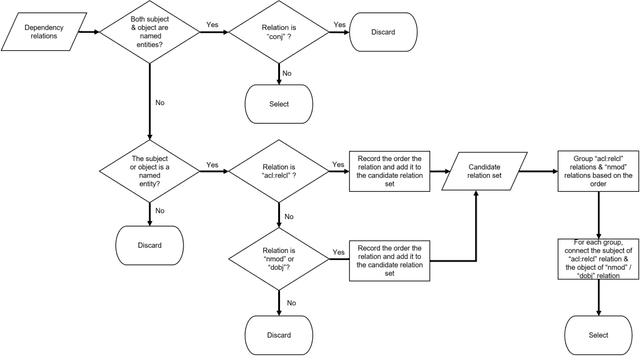
Abstract:With the ever-increasing scientific literature, there is a need on a natural language interface to bibliographic information retrieval systems to retrieve related information effectively. In this paper, we propose a natural language interface, NLI-GIBIR, to a graph-based bibliographic information retrieval system. In designing NLI-GIBIR, we developed a novel framework that can be applicable to graph-based bibliographic information retrieval systems. Our framework integrates algorithms/heuristics for interpreting and analyzing natural language bibliographic queries. NLI-GIBIR allows users to search for a variety of bibliographic data through natural language. A series of text- and linguistic-based techniques are used to analyze and answer natural language queries, including tokenization, named entity recognition, and syntactic analysis. We find that our framework can effectively represents and addresses complex bibliographic information needs. Thus, the contributions of this paper are as follows: First, to our knowledge, it is the first attempt to propose a natural language interface to graph-based bibliographic information retrieval. Second, we propose a novel customized natural language processing framework that integrates a few original algorithms/heuristics for interpreting and analyzing natural language bibliographic queries. Third, we show that the proposed framework and natural language interface provide a practical solution in building real-world natural language interface-based bibliographic information retrieval systems. Our experimental results show that the presented system can correctly answer 39 out of 40 example natural language queries with varying lengths and complexities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge