Catarina Botelho
CAMÕES: A Comprehensive Automatic Speech Recognition Benchmark for European Portuguese
Aug 27, 2025Abstract:Existing resources for Automatic Speech Recognition in Portuguese are mostly focused on Brazilian Portuguese, leaving European Portuguese (EP) and other varieties under-explored. To bridge this gap, we introduce CAM\~OES, the first open framework for EP and other Portuguese varieties. It consists of (1) a comprehensive evaluation benchmark, including 46h of EP test data spanning multiple domains; and (2) a collection of state-of-the-art models. For the latter, we consider multiple foundation models, evaluating their zero-shot and fine-tuned performances, as well as E-Branchformer models trained from scratch. A curated set of 425h of EP was used for both fine-tuning and training. Our results show comparable performance for EP between fine-tuned foundation models and the E-Branchformer. Furthermore, the best-performing models achieve relative improvements above 35% WER, compared to the strongest zero-shot foundation model, establishing a new state-of-the-art for EP and other varieties.
Tackling Cognitive Impairment Detection from Speech: A submission to the PROCESS Challenge
Dec 30, 2024



Abstract:This work describes our group's submission to the PROCESS Challenge 2024, with the goal of assessing cognitive decline through spontaneous speech, using three guided clinical tasks. This joint effort followed a holistic approach, encompassing both knowledge-based acoustic and text-based feature sets, as well as LLM-based macrolinguistic descriptors, pause-based acoustic biomarkers, and multiple neural representations (e.g., LongFormer, ECAPA-TDNN, and Trillson embeddings). Combining these feature sets with different classifiers resulted in a large pool of models, from which we selected those that provided the best balance between train, development, and individual class performance. Our results show that our best performing systems correspond to combinations of models that are complementary to each other, relying on acoustic and textual information from all three clinical tasks.
Unveiling Interpretability in Self-Supervised Speech Representations for Parkinson's Diagnosis
Dec 02, 2024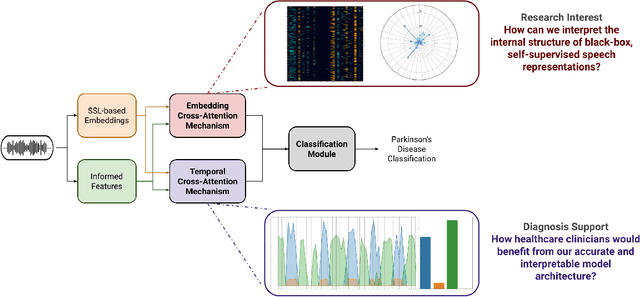
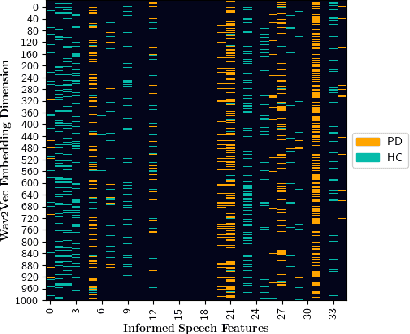
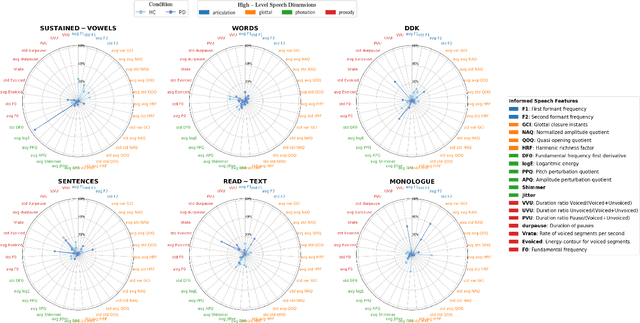
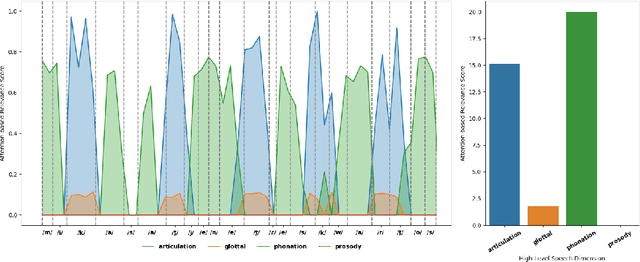
Abstract:Recent works in pathological speech analysis have increasingly relied on powerful self-supervised speech representations, leading to promising results. However, the complex, black-box nature of these embeddings and the limited research on their interpretability significantly restrict their adoption for clinical diagnosis. To address this gap, we propose a novel, interpretable framework specifically designed to support Parkinson's Disease (PD) diagnosis. Through the design of simple yet effective cross-attention mechanisms for both embedding- and temporal-level analysis, the proposed framework offers interpretability from two distinct but complementary perspectives. Experimental findings across five well-established speech benchmarks for PD detection demonstrate the framework's capability to identify meaningful speech patterns within self-supervised representations for a wide range of assessment tasks. Fine-grained temporal analyses further underscore its potential to enhance the interpretability of deep-learning pathological speech models, paving the way for the development of more transparent, trustworthy, and clinically applicable computer-assisted diagnosis systems in this domain. Moreover, in terms of classification accuracy, our method achieves results competitive with state-of-the-art approaches, while also demonstrating robustness in cross-lingual scenarios when applied to spontaneous speech production.
Speech as a Biomarker for Disease Detection
Sep 16, 2024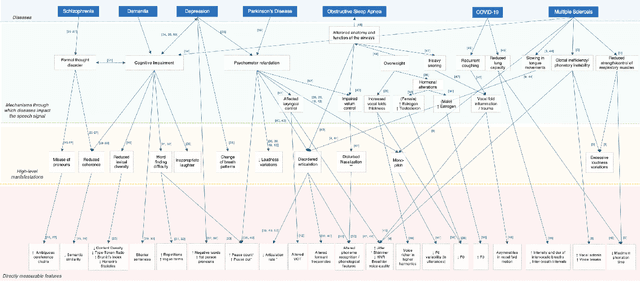
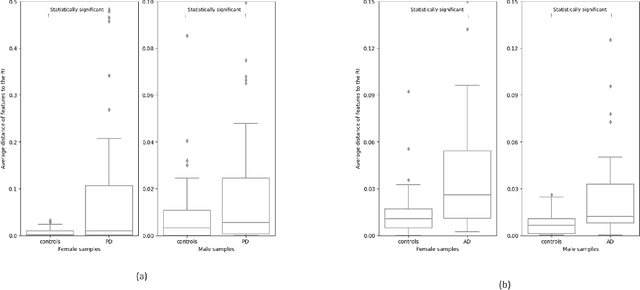

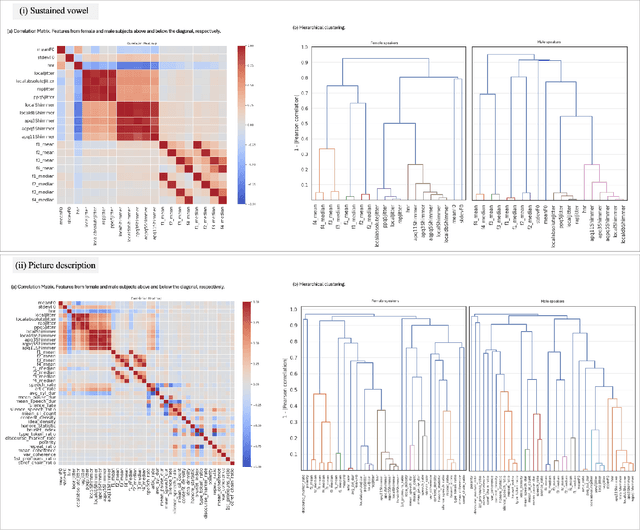
Abstract:Speech is a rich biomarker that encodes substantial information about the health of a speaker, and thus it has been proposed for the detection of numerous diseases, achieving promising results. However, questions remain about what the models trained for the automatic detection of these diseases are actually learning and the basis for their predictions, which can significantly impact patients' lives. This work advocates for an interpretable health model, suitable for detecting several diseases, motivated by the observation that speech-affecting disorders often have overlapping effects on speech signals. A framework is presented that first defines "reference speech" and then leverages this definition for disease detection. Reference speech is characterized through reference intervals, i.e., the typical values of clinically meaningful acoustic and linguistic features derived from a reference population. This novel approach in the field of speech as a biomarker is inspired by the use of reference intervals in clinical laboratory science. Deviations of new speakers from this reference model are quantified and used as input to detect Alzheimer's and Parkinson's disease. The classification strategy explored is based on Neural Additive Models, a type of glass-box neural network, which enables interpretability. The proposed framework for reference speech characterization and disease detection is designed to support the medical community by providing clinically meaningful explanations that can serve as a valuable second opinion.
Pathological speech detection using x-vector embeddings
Mar 03, 2020

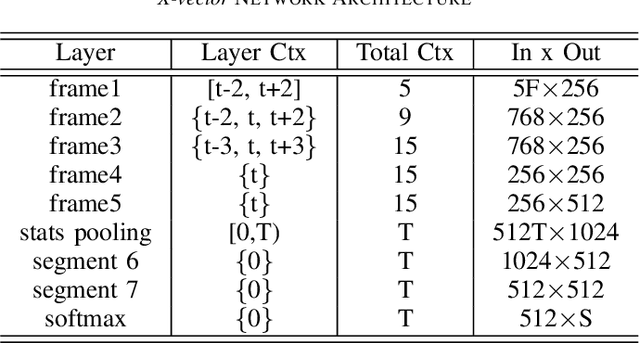

Abstract:The potential of speech as a non-invasive biomarker to assess a speaker's health has been repeatedly supported by the results of multiple works, for both physical and psychological conditions. Traditional systems for speech-based disease classification have focused on carefully designed knowledge-based features. However, these features may not represent the disease's full symptomatology, and may even overlook its more subtle manifestations. This has prompted researchers to move in the direction of general speaker representations that inherently model symptoms, such as Gaussian Supervectors, i-vectors and, x-vectors. In this work, we focus on the latter, to assess their applicability as a general feature extraction method to the detection of Parkinson's disease (PD) and obstructive sleep apnea (OSA). We test our approach against knowledge-based features and i-vectors, and report results for two European Portuguese corpora, for OSA and PD, as well as for an additional Spanish corpus for PD. Both x-vector and i-vector models were trained with an out-of-domain European Portuguese corpus. Our results show that x-vectors are able to perform better than knowledge-based features in same-language corpora. Moreover, while x-vectors performed similarly to i-vectors in matched conditions, they significantly outperform them when domain-mismatch occurs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge