Birgitta König-Ries
Explainability of Deep Learning-Based Plant Disease Classifiers Through Automated Concept Identification
Dec 10, 2024Abstract:While deep learning has significantly advanced automatic plant disease detection through image-based classification, improving model explainability remains crucial for reliable disease detection. In this study, we apply the Automated Concept-based Explanation (ACE) method to plant disease classification using the widely adopted InceptionV3 model and the PlantVillage dataset. ACE automatically identifies the visual concepts found in the image data and provides insights about the critical features influencing the model predictions. This approach reveals both effective disease-related patterns and incidental biases, such as those from background or lighting that can compromise model robustness. Through systematic experiments, ACE helped us to identify relevant features and pinpoint areas for targeted model improvement. Our findings demonstrate the potential of ACE to improve the explainability of plant disease classification based on deep learning, which is essential for producing transparent tools for plant disease management in agriculture.
GerPS-Compare: Comparing NER methods for legal norm analysis
Dec 03, 2024



Abstract:We apply NER to a particular sub-genre of legal texts in German: the genre of legal norms regulating administrative processes in public service administration. The analysis of such texts involves identifying stretches of text that instantiate one of ten classes identified by public service administration professionals. We investigate and compare three methods for performing Named Entity Recognition (NER) to detect these classes: a Rule-based system, deep discriminative models, and a deep generative model. Our results show that Deep Discriminative models outperform both the Rule-based system as well as the Deep Generative model, the latter two roughly performing equally well, outperforming each other in different classes. The main cause for this somewhat surprising result is arguably the fact that the classes used in the analysis are semantically and syntactically heterogeneous, in contrast to the classes used in more standard NER tasks. Deep Discriminative models appear to be better equipped for dealing with this heterogenerity than both generic LLMs and human linguists designing rule-based NER systems.
Harnessing multiple LLMs for Information Retrieval: A case study on Deep Learning methodologies in Biodiversity publications
Nov 14, 2024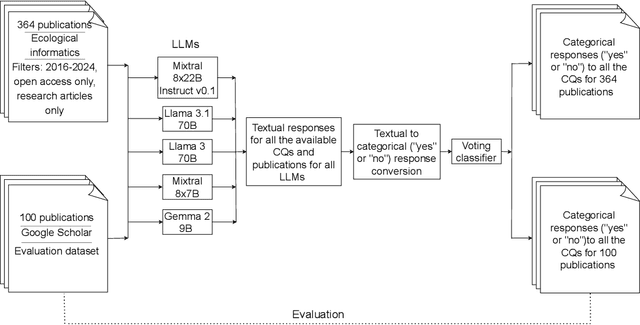
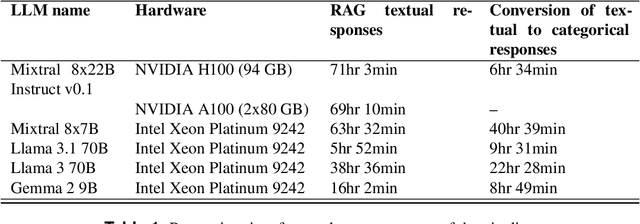

Abstract:Deep Learning (DL) techniques are increasingly applied in scientific studies across various domains to address complex research questions. However, the methodological details of these DL models are often hidden in the unstructured text. As a result, critical information about how these models are designed, trained, and evaluated is challenging to access and comprehend. To address this issue, in this work, we use five different open-source Large Language Models (LLMs): Llama-3 70B, Llama-3.1 70B, Mixtral-8x22B-Instruct-v0.1, Mixtral 8x7B, and Gemma 2 9B in combination with Retrieval-Augmented Generation (RAG) approach to extract and process DL methodological details from scientific publications automatically. We built a voting classifier from the outputs of five LLMs to accurately report DL methodological information. We tested our approach using biodiversity publications, building upon our previous research. To validate our pipeline, we employed two datasets of DL-related biodiversity publications: a curated set of 100 publications from our prior work and a set of 364 publications from the Ecological Informatics journal. Our results demonstrate that the multi-LLM, RAG-assisted pipeline enhances the retrieval of DL methodological information, achieving an accuracy of 69.5% (417 out of 600 comparisons) based solely on textual content from publications. This performance was assessed against human annotators who had access to code, figures, tables, and other supplementary information. Although demonstrated in biodiversity, our methodology is not limited to this field; it can be applied across other scientific domains where detailed methodological reporting is essential for advancing knowledge and ensuring reproducibility. This study presents a scalable and reliable approach for automating information extraction, facilitating better reproducibility and knowledge transfer across studies.
Enhancing Explainability in Multimodal Large Language Models Using Ontological Context
Sep 27, 2024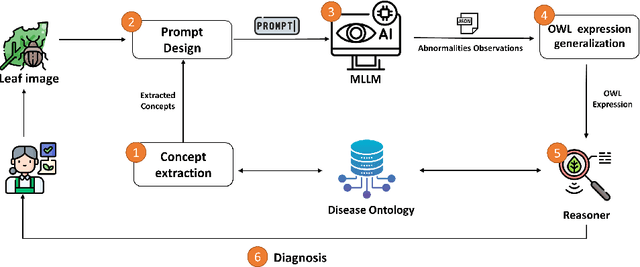



Abstract:Recently, there has been a growing interest in Multimodal Large Language Models (MLLMs) due to their remarkable potential in various tasks integrating different modalities, such as image and text, as well as applications such as image captioning and visual question answering. However, such models still face challenges in accurately captioning and interpreting specific visual concepts and classes, particularly in domain-specific applications. We argue that integrating domain knowledge in the form of an ontology can significantly address these issues. In this work, as a proof of concept, we propose a new framework that combines ontology with MLLMs to classify images of plant diseases. Our method uses concepts about plant diseases from an existing disease ontology to query MLLMs and extract relevant visual concepts from images. Then, we use the reasoning capabilities of the ontology to classify the disease according to the identified concepts. Ensuring that the model accurately uses the concepts describing the disease is crucial in domain-specific applications. By employing an ontology, we can assist in verifying this alignment. Additionally, using the ontology's inference capabilities increases transparency, explainability, and trust in the decision-making process while serving as a judge by checking if the annotations of the concepts by MLLMs are aligned with those in the ontology and displaying the rationales behind their errors. Our framework offers a new direction for synergizing ontologies and MLLMs, supported by an empirical study using different well-known MLLMs.
Evaluating the method reproducibility of deep learning models in the biodiversity domain
Jul 10, 2024Abstract:Artificial Intelligence (AI) is revolutionizing biodiversity research by enabling advanced data analysis, species identification, and habitats monitoring, thereby enhancing conservation efforts. Ensuring reproducibility in AI-driven biodiversity research is crucial for fostering transparency, verifying results, and promoting the credibility of ecological findings.This study investigates the reproducibility of deep learning (DL) methods within the biodiversity domain. We design a methodology for evaluating the reproducibility of biodiversity-related publications that employ DL techniques across three stages. We define ten variables essential for method reproducibility, divided into four categories: resource requirements, methodological information, uncontrolled randomness, and statistical considerations. These categories subsequently serve as the basis for defining different levels of reproducibility. We manually extract the availability of these variables from a curated dataset comprising 61 publications identified using the keywords provided by biodiversity experts. Our study shows that the dataset is shared in 47% of the publications; however, a significant number of the publications lack comprehensive information on deep learning methods, including details regarding randomness.
From human experts to machines: An LLM supported approach to ontology and knowledge graph construction
Mar 13, 2024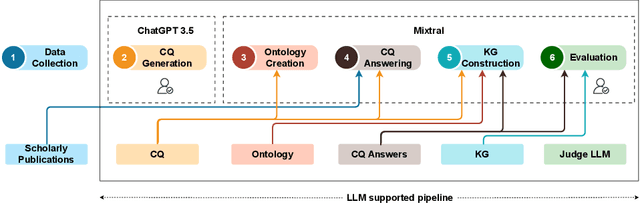
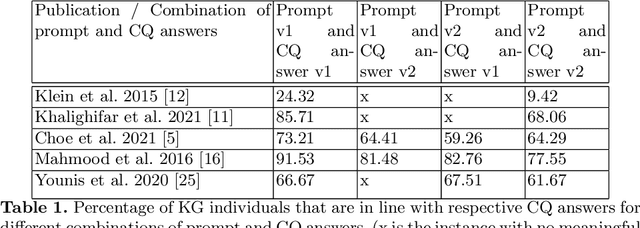
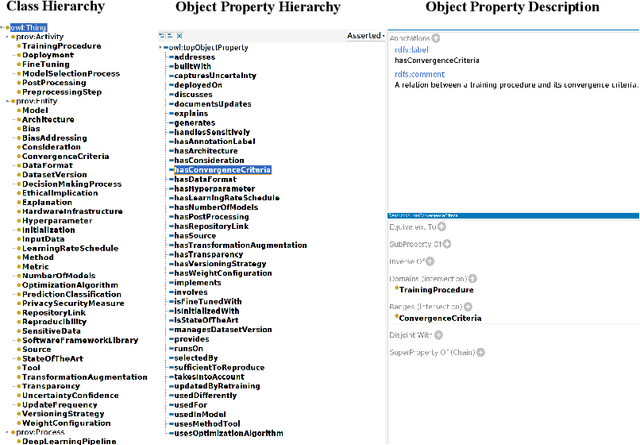
Abstract:The conventional process of building Ontologies and Knowledge Graphs (KGs) heavily relies on human domain experts to define entities and relationship types, establish hierarchies, maintain relevance to the domain, fill the ABox (or populate with instances), and ensure data quality (including amongst others accuracy and completeness). On the other hand, Large Language Models (LLMs) have recently gained popularity for their ability to understand and generate human-like natural language, offering promising ways to automate aspects of this process. This work explores the (semi-)automatic construction of KGs facilitated by open-source LLMs. Our pipeline involves formulating competency questions (CQs), developing an ontology (TBox) based on these CQs, constructing KGs using the developed ontology, and evaluating the resultant KG with minimal to no involvement of human experts. We showcase the feasibility of our semi-automated pipeline by creating a KG on deep learning methodologies by exploiting scholarly publications. To evaluate the answers generated via Retrieval-Augmented-Generation (RAG) as well as the KG concepts automatically extracted using LLMs, we design a judge LLM, which rates the generated content based on ground truth. Our findings suggest that employing LLMs could potentially reduce the human effort involved in the construction of KGs, although a human-in-the-loop approach is recommended to evaluate automatically generated KGs.
Concept explainability for plant diseases classification
Sep 15, 2023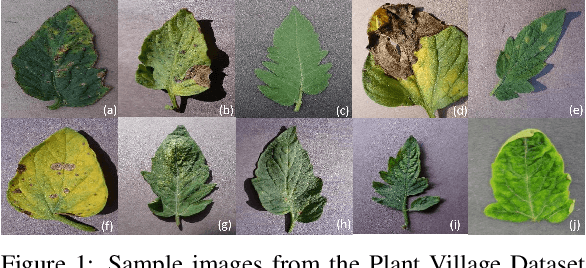

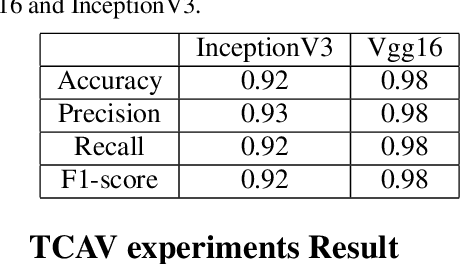
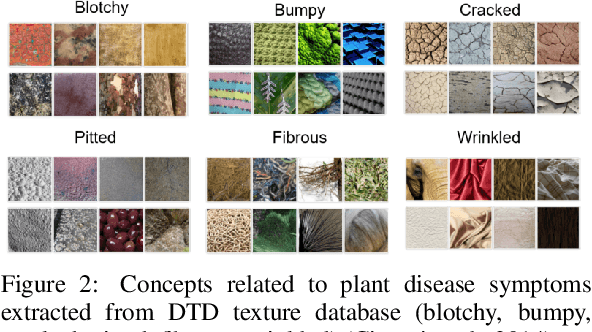
Abstract:Plant diseases remain a considerable threat to food security and agricultural sustainability. Rapid and early identification of these diseases has become a significant concern motivating several studies to rely on the increasing global digitalization and the recent advances in computer vision based on deep learning. In fact, plant disease classification based on deep convolutional neural networks has shown impressive performance. However, these methods have yet to be adopted globally due to concerns regarding their robustness, transparency, and the lack of explainability compared with their human experts counterparts. Methods such as saliency-based approaches associating the network output to perturbations of the input pixels have been proposed to give insights into these algorithms. Still, they are not easily comprehensible and not intuitive for human users and are threatened by bias. In this work, we deploy a method called Testing with Concept Activation Vectors (TCAV) that shifts the focus from pixels to user-defined concepts. To the best of our knowledge, our paper is the first to employ this method in the field of plant disease classification. Important concepts such as color, texture and disease related concepts were analyzed. The results suggest that concept-based explanation methods can significantly benefit automated plant disease identification.
* Accepted at VISAPP 2023
Reproducible Domain-Specific Knowledge Graphs in the Life Sciences: a Systematic Literature Review
Sep 15, 2023Abstract:Knowledge graphs (KGs) are widely used for representing and organizing structured knowledge in diverse domains. However, the creation and upkeep of KGs pose substantial challenges. Developing a KG demands extensive expertise in data modeling, ontology design, and data curation. Furthermore, KGs are dynamic, requiring continuous updates and quality control to ensure accuracy and relevance. These intricacies contribute to the considerable effort required for their development and maintenance. One critical dimension of KGs that warrants attention is reproducibility. The ability to replicate and validate KGs is fundamental for ensuring the trustworthiness and sustainability of the knowledge they represent. Reproducible KGs not only support open science by allowing others to build upon existing knowledge but also enhance transparency and reliability in disseminating information. Despite the growing number of domain-specific KGs, a comprehensive analysis concerning their reproducibility has been lacking. This paper addresses this gap by offering a general overview of domain-specific KGs and comparing them based on various reproducibility criteria. Our study over 19 different domains shows only eight out of 250 domain-specific KGs (3.2%) provide publicly available source code. Among these, only one system could successfully pass our reproducibility assessment (14.3%). These findings highlight the challenges and gaps in achieving reproducibility across domain-specific KGs. Our finding that only 0.4% of published domain-specific KGs are reproducible shows a clear need for further research and a shift in cultural practices.
Machine Learning Pipelines: Provenance, Reproducibility and FAIR Data Principles
Jun 22, 2020Abstract:Machine learning (ML) is an increasingly important scientific tool supporting decision making and knowledge generation in numerous fields. With this, it also becomes more and more important that the results of ML experiments are reproducible. Unfortunately, that often is not the case. Rather, ML, similar to many other disciplines, faces a reproducibility crisis. In this paper, we describe our goals and initial steps in supporting the end-to-end reproducibility of ML pipelines. We investigate which factors beyond the availability of source code and datasets influence reproducibility of ML experiments. We propose ways to apply FAIR data practices to ML workflows. We present our preliminary results on the role of our tool, ProvBook, in capturing and comparing provenance of ML experiments and their reproducibility using Jupyter Notebooks.
Towards Building Knowledge by Merging Multiple Ontologies with CoMerger: A Partitioning-based Approach
May 06, 2020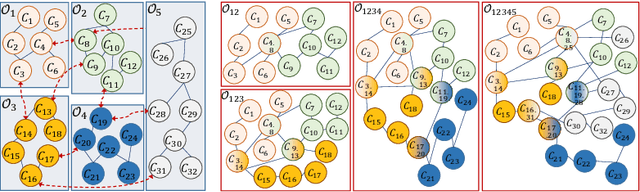
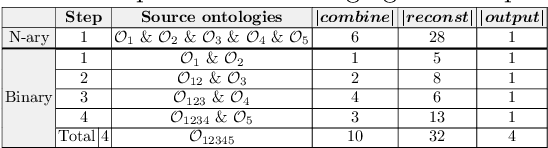
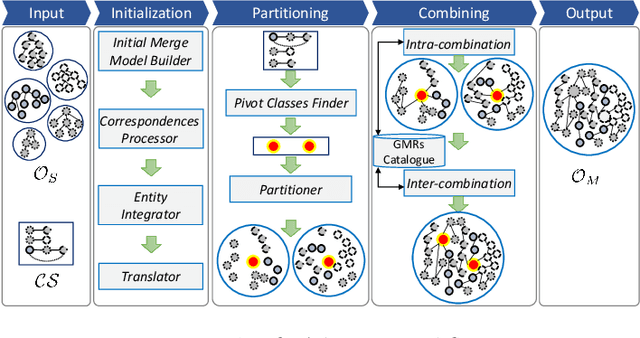

Abstract:Ontologies are the prime way of organizing data in the Semantic Web. Often, it is necessary to combine several, independently developed ontologies to obtain a knowledge graph fully representing a domain of interest. The complementarity of existing ontologies can be leveraged by merging them. Existing approaches for ontology merging mostly implement a binary merge. However, with the growing number and size of relevant ontologies across domains, scalability becomes a central challenge. A multi-ontology merging technique offers a potential solution to this problem. We present CoMerger, a scalable multiple ontologies merging method. For efficient processing, rather than successively merging complete ontologies pairwise, we group related concepts across ontologies into partitions and merge first within and then across those partitions. The experimental results on well-known datasets confirm the feasibility of our approach and demonstrate its superiority over binary strategies. A prototypical implementation is freely accessible through a live web portal.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge