Harnessing multiple LLMs for Information Retrieval: A case study on Deep Learning methodologies in Biodiversity publications
Paper and Code
Nov 14, 2024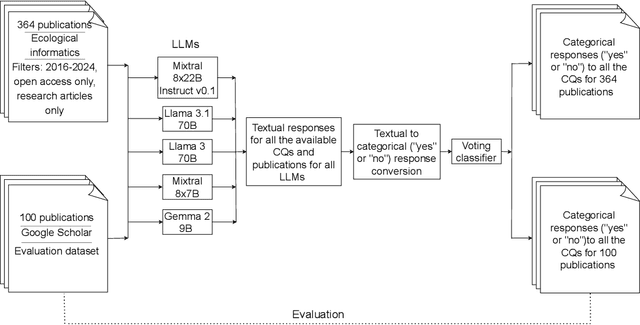
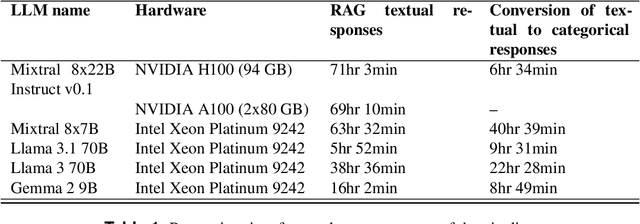

Deep Learning (DL) techniques are increasingly applied in scientific studies across various domains to address complex research questions. However, the methodological details of these DL models are often hidden in the unstructured text. As a result, critical information about how these models are designed, trained, and evaluated is challenging to access and comprehend. To address this issue, in this work, we use five different open-source Large Language Models (LLMs): Llama-3 70B, Llama-3.1 70B, Mixtral-8x22B-Instruct-v0.1, Mixtral 8x7B, and Gemma 2 9B in combination with Retrieval-Augmented Generation (RAG) approach to extract and process DL methodological details from scientific publications automatically. We built a voting classifier from the outputs of five LLMs to accurately report DL methodological information. We tested our approach using biodiversity publications, building upon our previous research. To validate our pipeline, we employed two datasets of DL-related biodiversity publications: a curated set of 100 publications from our prior work and a set of 364 publications from the Ecological Informatics journal. Our results demonstrate that the multi-LLM, RAG-assisted pipeline enhances the retrieval of DL methodological information, achieving an accuracy of 69.5% (417 out of 600 comparisons) based solely on textual content from publications. This performance was assessed against human annotators who had access to code, figures, tables, and other supplementary information. Although demonstrated in biodiversity, our methodology is not limited to this field; it can be applied across other scientific domains where detailed methodological reporting is essential for advancing knowledge and ensuring reproducibility. This study presents a scalable and reliable approach for automating information extraction, facilitating better reproducibility and knowledge transfer across studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge