Jihen Amara
Explainability of Deep Learning-Based Plant Disease Classifiers Through Automated Concept Identification
Dec 10, 2024Abstract:While deep learning has significantly advanced automatic plant disease detection through image-based classification, improving model explainability remains crucial for reliable disease detection. In this study, we apply the Automated Concept-based Explanation (ACE) method to plant disease classification using the widely adopted InceptionV3 model and the PlantVillage dataset. ACE automatically identifies the visual concepts found in the image data and provides insights about the critical features influencing the model predictions. This approach reveals both effective disease-related patterns and incidental biases, such as those from background or lighting that can compromise model robustness. Through systematic experiments, ACE helped us to identify relevant features and pinpoint areas for targeted model improvement. Our findings demonstrate the potential of ACE to improve the explainability of plant disease classification based on deep learning, which is essential for producing transparent tools for plant disease management in agriculture.
Enhancing Explainability in Multimodal Large Language Models Using Ontological Context
Sep 27, 2024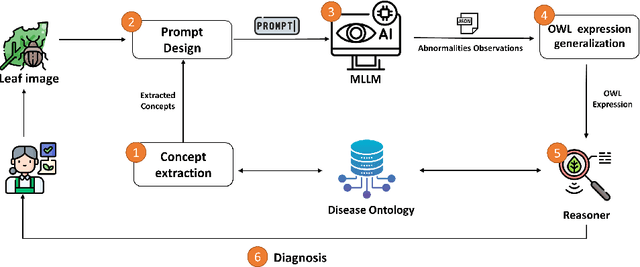



Abstract:Recently, there has been a growing interest in Multimodal Large Language Models (MLLMs) due to their remarkable potential in various tasks integrating different modalities, such as image and text, as well as applications such as image captioning and visual question answering. However, such models still face challenges in accurately captioning and interpreting specific visual concepts and classes, particularly in domain-specific applications. We argue that integrating domain knowledge in the form of an ontology can significantly address these issues. In this work, as a proof of concept, we propose a new framework that combines ontology with MLLMs to classify images of plant diseases. Our method uses concepts about plant diseases from an existing disease ontology to query MLLMs and extract relevant visual concepts from images. Then, we use the reasoning capabilities of the ontology to classify the disease according to the identified concepts. Ensuring that the model accurately uses the concepts describing the disease is crucial in domain-specific applications. By employing an ontology, we can assist in verifying this alignment. Additionally, using the ontology's inference capabilities increases transparency, explainability, and trust in the decision-making process while serving as a judge by checking if the annotations of the concepts by MLLMs are aligned with those in the ontology and displaying the rationales behind their errors. Our framework offers a new direction for synergizing ontologies and MLLMs, supported by an empirical study using different well-known MLLMs.
Concept explainability for plant diseases classification
Sep 15, 2023

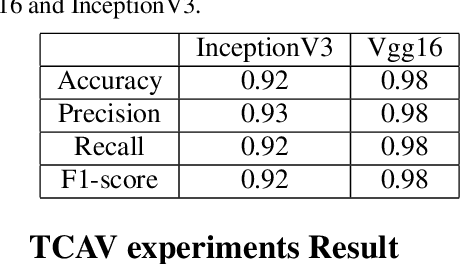
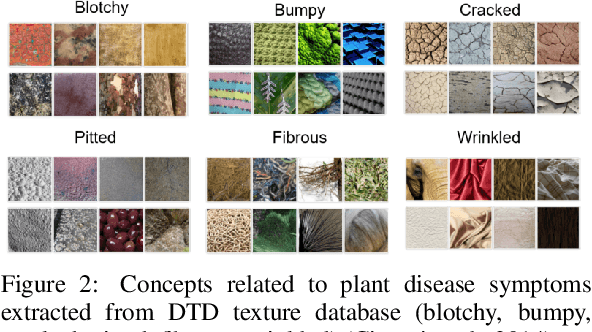
Abstract:Plant diseases remain a considerable threat to food security and agricultural sustainability. Rapid and early identification of these diseases has become a significant concern motivating several studies to rely on the increasing global digitalization and the recent advances in computer vision based on deep learning. In fact, plant disease classification based on deep convolutional neural networks has shown impressive performance. However, these methods have yet to be adopted globally due to concerns regarding their robustness, transparency, and the lack of explainability compared with their human experts counterparts. Methods such as saliency-based approaches associating the network output to perturbations of the input pixels have been proposed to give insights into these algorithms. Still, they are not easily comprehensible and not intuitive for human users and are threatened by bias. In this work, we deploy a method called Testing with Concept Activation Vectors (TCAV) that shifts the focus from pixels to user-defined concepts. To the best of our knowledge, our paper is the first to employ this method in the field of plant disease classification. Important concepts such as color, texture and disease related concepts were analyzed. The results suggest that concept-based explanation methods can significantly benefit automated plant disease identification.
* Accepted at VISAPP 2023
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge