Betty Chan
Gemini: A Family of Highly Capable Multimodal Models
Dec 19, 2023Abstract:This report introduces a new family of multimodal models, Gemini, that exhibit remarkable capabilities across image, audio, video, and text understanding. The Gemini family consists of Ultra, Pro, and Nano sizes, suitable for applications ranging from complex reasoning tasks to on-device memory-constrained use-cases. Evaluation on a broad range of benchmarks shows that our most-capable Gemini Ultra model advances the state of the art in 30 of 32 of these benchmarks - notably being the first model to achieve human-expert performance on the well-studied exam benchmark MMLU, and improving the state of the art in every one of the 20 multimodal benchmarks we examined. We believe that the new capabilities of Gemini models in cross-modal reasoning and language understanding will enable a wide variety of use cases and we discuss our approach toward deploying them responsibly to users.
Machine learning on DNA-encoded libraries: A new paradigm for hit-finding
Jan 31, 2020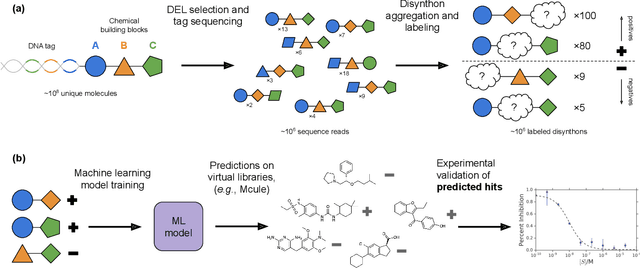
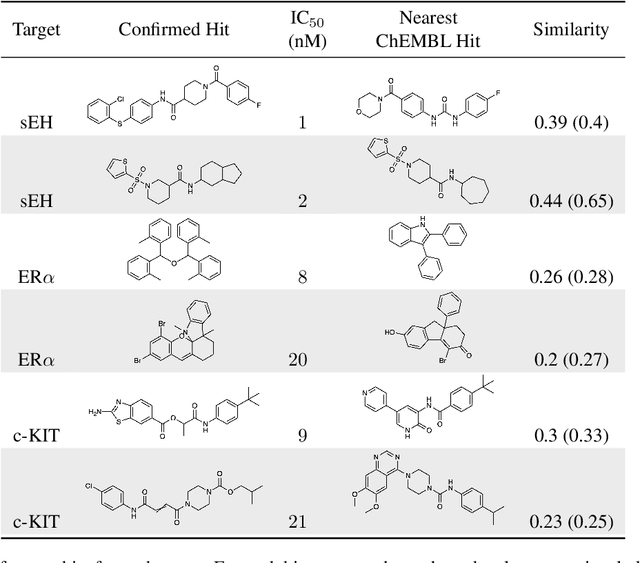
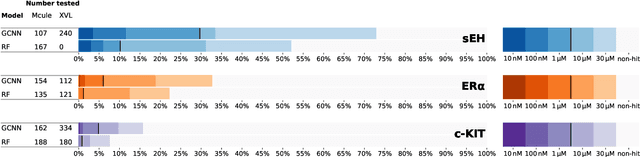
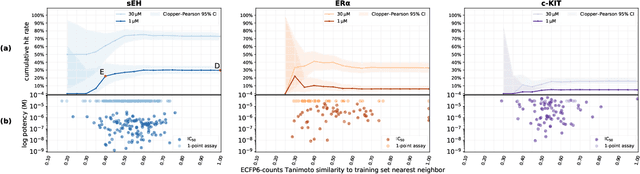
Abstract:DNA-encoded small molecule libraries (DELs) have enabled discovery of novel inhibitors for many distinct protein targets of therapeutic value through screening of libraries with up to billions of unique small molecules. We demonstrate a new approach applying machine learning to DEL selection data by identifying active molecules from a large commercial collection and a virtual library of easily synthesizable compounds. We train models using only DEL selection data and apply automated or automatable filters with chemist review restricted to the removal of molecules with potential for instability or reactivity. We validate this approach with a large prospective study (nearly 2000 compounds tested) across three diverse protein targets: sEH (a hydrolase), ER{\alpha} (a nuclear receptor), and c-KIT (a kinase). The approach is effective, with an overall hit rate of {\sim}30% at 30 {\textmu}M and discovery of potent compounds (IC50 <10 nM) for every target. The model makes useful predictions even for molecules dissimilar to the original DEL and the compounds identified are diverse, predominantly drug-like, and different from known ligands. Collectively, the quality and quantity of DEL selection data; the power of modern machine learning methods; and access to large, inexpensive, commercially-available libraries creates a powerful new approach for hit finding.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge