Attila Losonczy
A zero-inflated gamma model for deconvolved calcium imaging traces
Jun 05, 2020Abstract:Calcium imaging is a critical tool for measuring the activity of large neural populations. Much effort has been devoted to developing "pre-processing" tools for calcium video data, addressing the important issues of e.g., motion correction, denoising, compression, demixing, and deconvolution. However, statistical modeling of deconvolved calcium signals (i.e., the estimated activity extracted by a pre-processing pipeline) is just as critical for interpreting calcium measurements, and for incorporating these observations into downstream probabilistic encoding and decoding models. Surprisingly, these issues have to date received significantly less attention. In this work we examine the statistical properties of the deconvolved activity estimates, and compare probabilistic models for these random signals. In particular, we propose a zero-inflated gamma (ZIG) model, which characterizes the calcium responses as a mixture of a gamma distribution and a point mass that serves to model zero responses. We apply the resulting models to neural encoding and decoding problems. We find that the ZIG model outperforms simpler models (e.g., Poisson or Bernoulli models) in the context of both simulated and real neural data, and can therefore play a useful role in bridging calcium imaging analysis methods with tools for analyzing activity in large neural populations.
* Accepted for publication in Neurons, Behavior, Data analysis, and Theory
Artificial neural networks in action for an automated cell-type classification of biological neural networks
Nov 22, 2019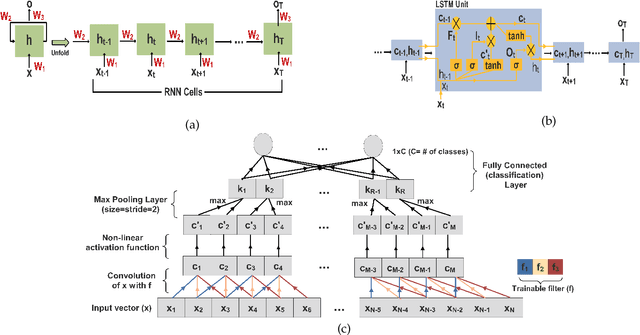
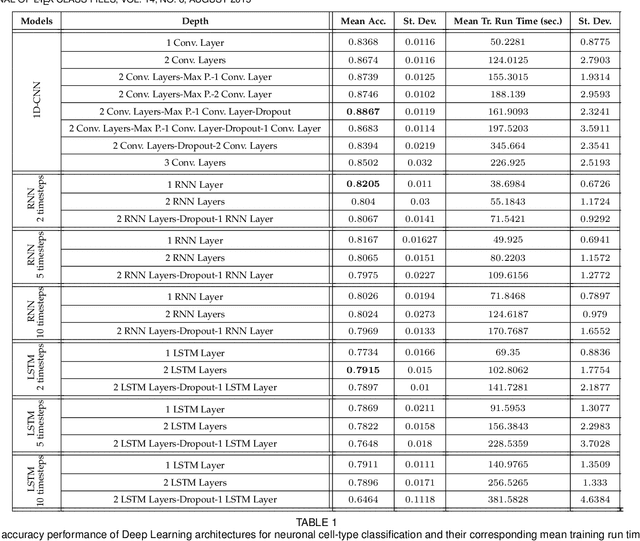
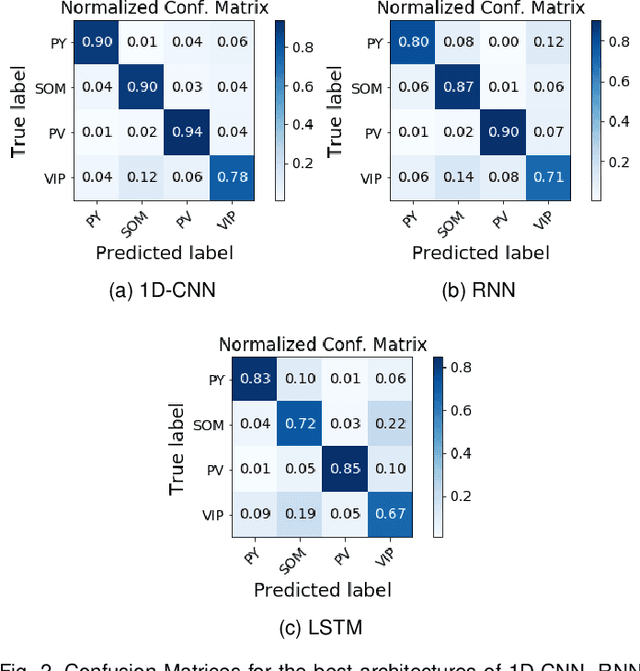
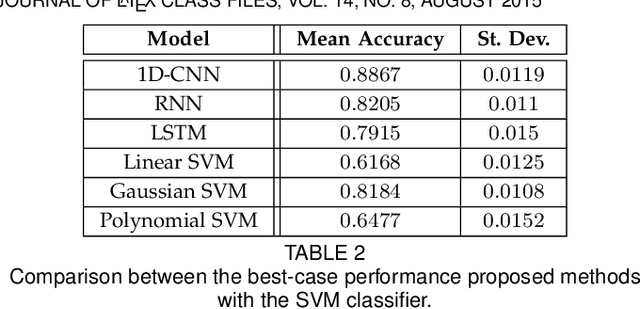
Abstract:In this work we address the problem of neuronal cell-type classification, and we employ a real-world dataset of raw neuronal activity measurements obtained with calcium imaging techniques. While neuronal cell-type classification is a crucial step in understanding the function of neuronal circuits, and thus a systematic classification of neurons is much needed, it still remains a challenge. In recent years, several approaches have been employed for a reliable neuronal cell-type recognition, such as immunohistochemical (IHC) analysis and feature extraction algorithms based on several characteristics of neuronal cells. These methods, however, demand a lot of human intervention and observation, they are time-consuming and regarding the feature extraction algorithms it is not clear or obvious what are the best features that define a neuronal cell class. In this work we examine three different deep learning models aiming at an automated neuronal cell-type classification and compare their performance. Experimental analysis demonstrates the efficacy and potent capabilities for each one of the proposed schemes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge