Spyridon Chavlis
Dendrites endow artificial neural networks with accurate, robust and parameter-efficient learning
Apr 04, 2024Abstract:Artificial neural networks (ANNs) are at the core of most Deep learning (DL) algorithms that successfully tackle complex problems like image recognition, autonomous driving, and natural language processing. However, unlike biological brains who tackle similar problems in a very efficient manner, DL algorithms require a large number of trainable parameters, making them energy-intensive and prone to overfitting. Here, we show that a new ANN architecture that incorporates the structured connectivity and restricted sampling properties of biological dendrites counteracts these limitations. We find that dendritic ANNs are more robust to overfitting and outperform traditional ANNs on several image classification tasks while using significantly fewer trainable parameters. This is achieved through the adoption of a different learning strategy, whereby most of the nodes respond to several classes, unlike classical ANNs that strive for class-specificity. These findings suggest that the incorporation of dendrites can make learning in ANNs precise, resilient, and parameter-efficient and shed new light on how biological features can impact the learning strategies of ANNs.
Dendritic Self-Organizing Maps for Continual Learning
Oct 18, 2021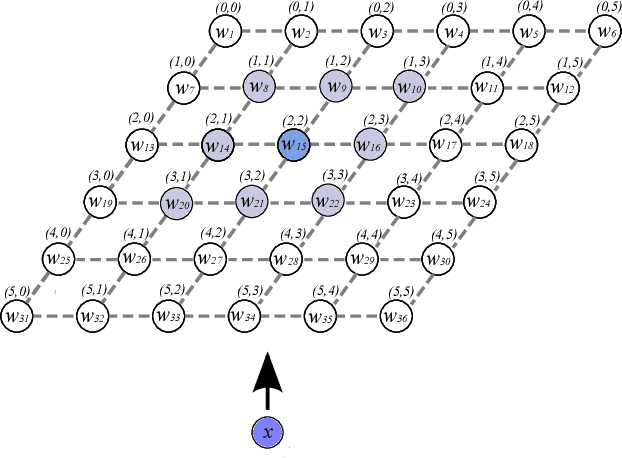

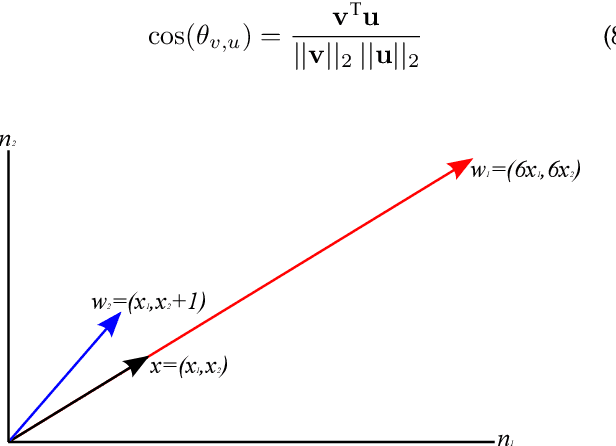
Abstract:Current deep learning architectures show remarkable performance when trained in large-scale, controlled datasets. However, the predictive ability of these architectures significantly decreases when learning new classes incrementally. This is due to their inclination to forget the knowledge acquired from previously seen data, a phenomenon termed catastrophic-forgetting. On the other hand, Self-Organizing Maps (SOMs) can model the input space utilizing constrained k-means and thus maintain past knowledge. Here, we propose a novel algorithm inspired by biological neurons, termed Dendritic-Self-Organizing Map (DendSOM). DendSOM consists of a single layer of SOMs, which extract patterns from specific regions of the input space accompanied by a set of hit matrices, one per SOM, which estimate the association between units and labels. The best-matching unit of an input pattern is selected using the maximum cosine similarity rule, while the point-wise mutual information is employed for class inference. DendSOM performs unsupervised feature extraction as it does not use labels for targeted updating of the weights. It outperforms classical SOMs and several state-of-the-art continual learning algorithms on benchmark datasets, such as the Split-MNIST and Split-CIFAR-10. We propose that the incorporation of neuronal properties in SOMs may help remedy catastrophic forgetting.
Artificial neural networks in action for an automated cell-type classification of biological neural networks
Nov 22, 2019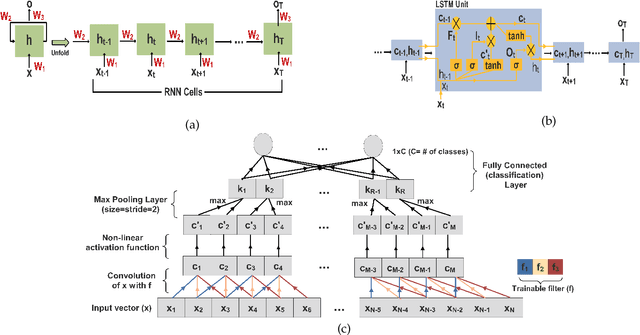
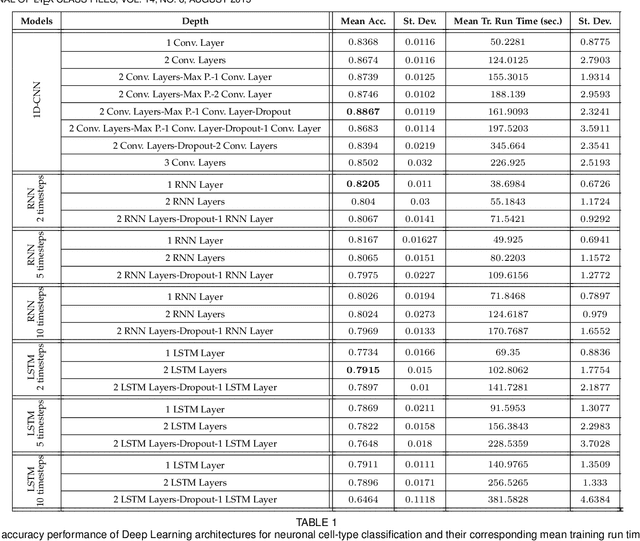
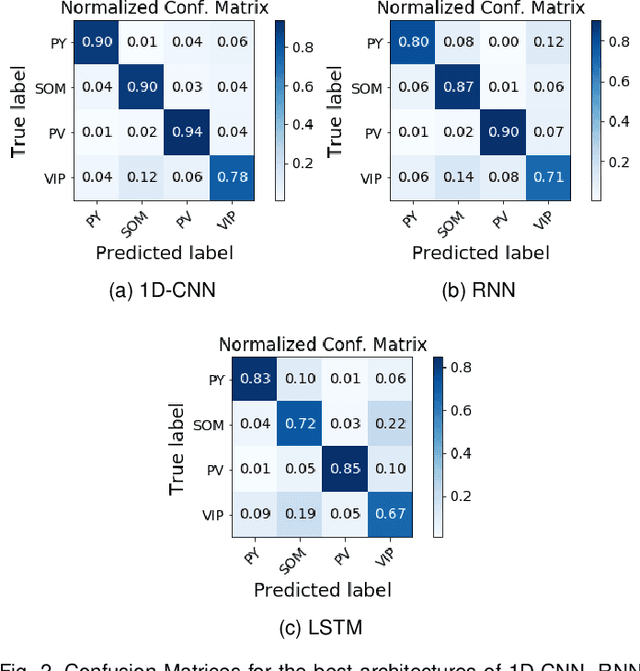
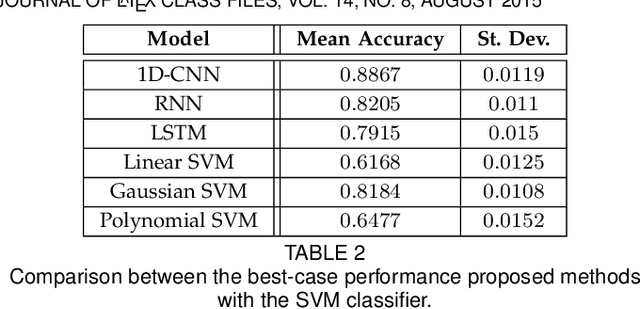
Abstract:In this work we address the problem of neuronal cell-type classification, and we employ a real-world dataset of raw neuronal activity measurements obtained with calcium imaging techniques. While neuronal cell-type classification is a crucial step in understanding the function of neuronal circuits, and thus a systematic classification of neurons is much needed, it still remains a challenge. In recent years, several approaches have been employed for a reliable neuronal cell-type recognition, such as immunohistochemical (IHC) analysis and feature extraction algorithms based on several characteristics of neuronal cells. These methods, however, demand a lot of human intervention and observation, they are time-consuming and regarding the feature extraction algorithms it is not clear or obvious what are the best features that define a neuronal cell class. In this work we examine three different deep learning models aiming at an automated neuronal cell-type classification and compare their performance. Experimental analysis demonstrates the efficacy and potent capabilities for each one of the proposed schemes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge