Andrew J. Saykin
Multidimensional representations in late-life depression: convergence in neuroimaging, cognition, clinical symptomatology and genetics
Oct 25, 2021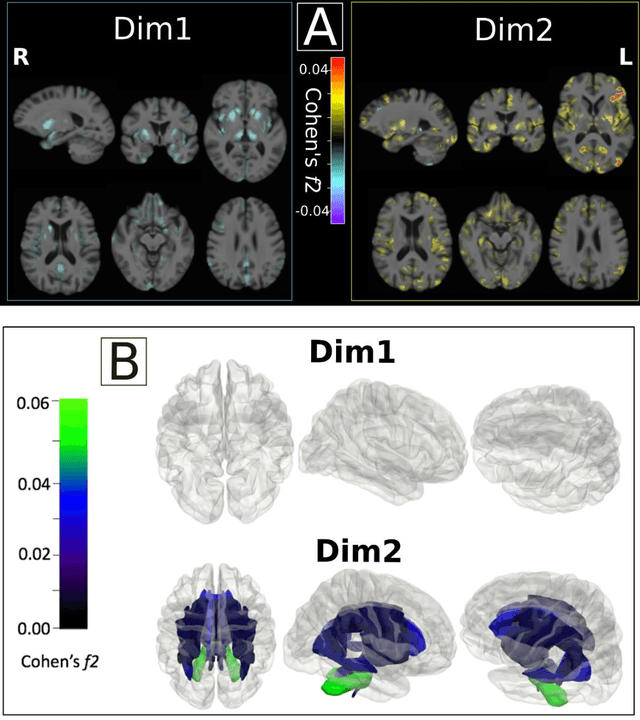
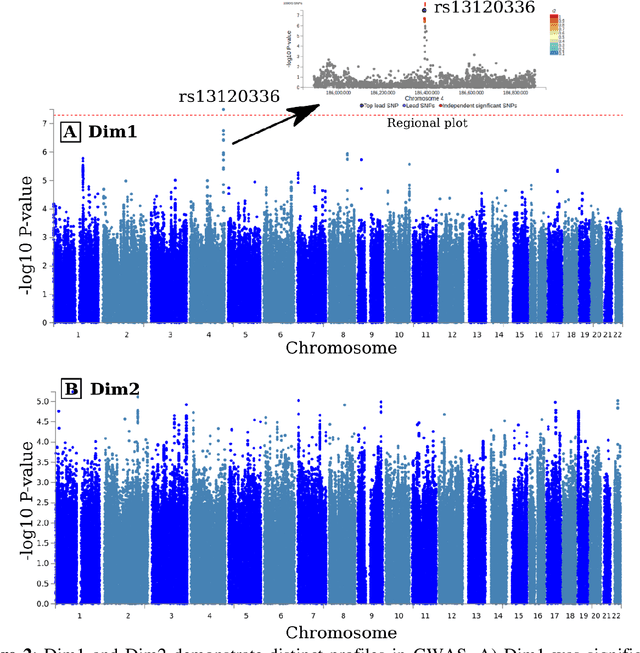
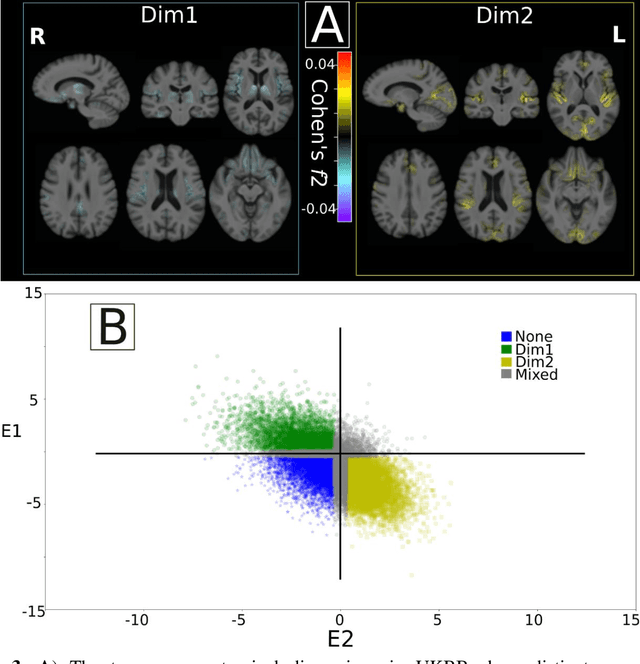
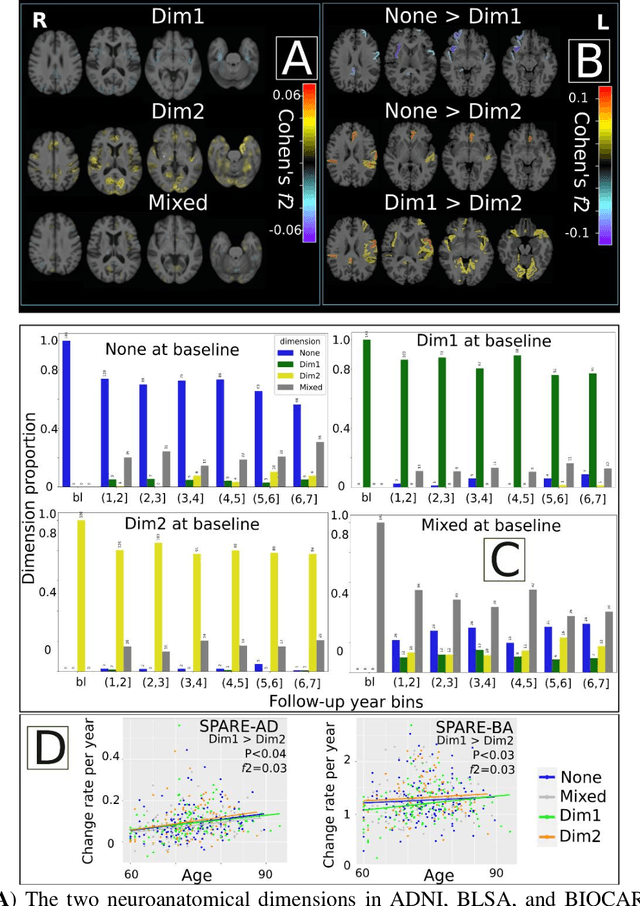
Abstract:Late-life depression (LLD) is characterized by considerable heterogeneity in clinical manifestation. Unraveling such heterogeneity would aid in elucidating etiological mechanisms and pave the road to precision and individualized medicine. We sought to delineate, cross-sectionally and longitudinally, disease-related heterogeneity in LLD linked to neuroanatomy, cognitive functioning, clinical symptomatology, and genetic profiles. Multimodal data from a multicentre sample (N=996) were analyzed. A semi-supervised clustering method (HYDRA) was applied to regional grey matter (GM) brain volumes to derive dimensional representations. Two dimensions were identified, which accounted for the LLD-related heterogeneity in voxel-wise GM maps, white matter (WM) fractional anisotropy (FA), neurocognitive functioning, clinical phenotype, and genetics. Dimension one (Dim1) demonstrated relatively preserved brain anatomy without WM disruptions relative to healthy controls. In contrast, dimension two (Dim2) showed widespread brain atrophy and WM integrity disruptions, along with cognitive impairment and higher depression severity. Moreover, one de novo independent genetic variant (rs13120336) was significantly associated with Dim 1 but not with Dim 2. Notably, the two dimensions demonstrated significant SNP-based heritability of 18-27% within the general population (N=12,518 in UKBB). Lastly, in a subset of individuals having longitudinal measurements, Dim2 demonstrated a more rapid longitudinal decrease in GM and brain age, and was more likely to progress to Alzheimers disease, compared to Dim1 (N=1,413 participants and 7,225 scans from ADNI, BLSA, and BIOCARD datasets).
Deep Learning in Alzheimer's disease: Diagnostic Classification and Prognostic Prediction using Neuroimaging Data
May 06, 2019
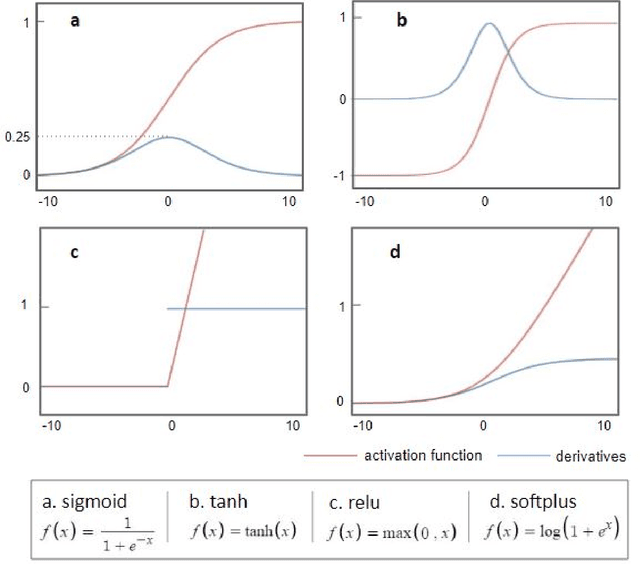
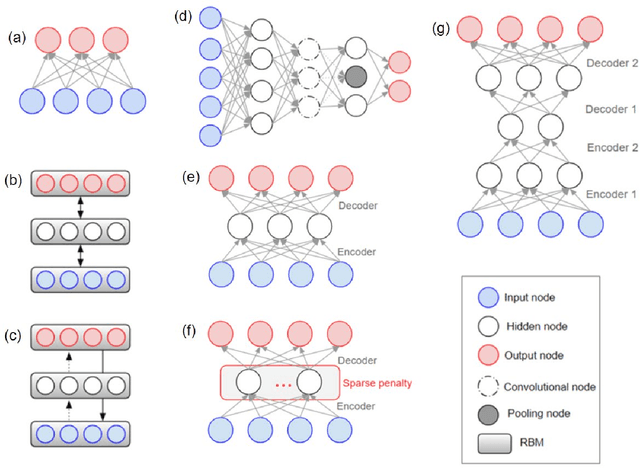
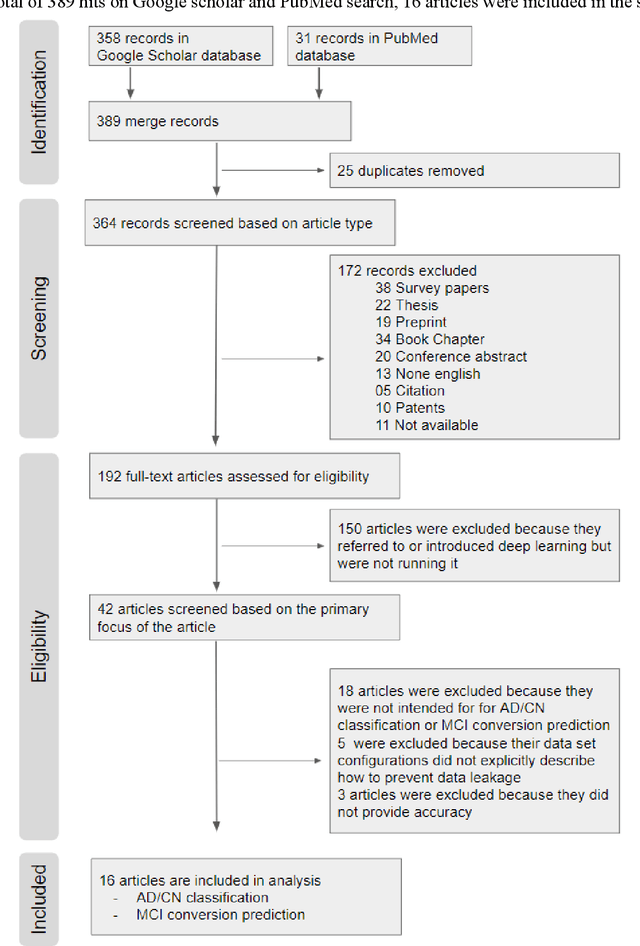
Abstract:The application of deep learning to early detection and automated classification of Alzheimer's disease (AD) has recently gained considerable attention as rapid progress in neuroimaging techniques has generated large-scale multimodal neuroimaging data. Here we systematically reviewed publications, where deep learning approaches and neuroimaging data were used for diagnostic classification of AD. A PubMed and google scholar search was performed to find deep learning papers for AD published between January 2013 and July 2018, which were reviewed, evaluated, and classified by algorithms and neuroimaging types, and findings were summarized. The diagnostic classification of AD using deep learning approaches and neuroimaging data was examined in 16 studies. The approach to combine traditional machine learning for classification and stacked auto-encoder (SAE) for feature selection has produced accuracies of up to 98.8% for AD classification and 83.7% for prediction of conversion from mild cognitive impairment (MCI), a prodromal stage of AD, to AD. Deep learning approaches such as convolutional neural network (CNN) or recurrent neural network (RNN) using neuroimaging data without preprocessing for feature selection have yielded accuracies of up to 96.0% for AD classification and 84.2% for MCI conversion prediction. Furthermore, the best classification performance was obtained when multimodal neuroimaging data as well as fluid biomarkers were integrated. Deep learning approaches without preprocessing neuroimaging data for feature selection, a major bottleneck of traditional machining learning in high-dimensional data, continue to improve their performance and to show great promise in the diagnostic classification of AD using multimodal neuroimaging data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge