Taeho Jo
Deep Learning in Alzheimer's disease: Diagnostic Classification and Prognostic Prediction using Neuroimaging Data
May 06, 2019
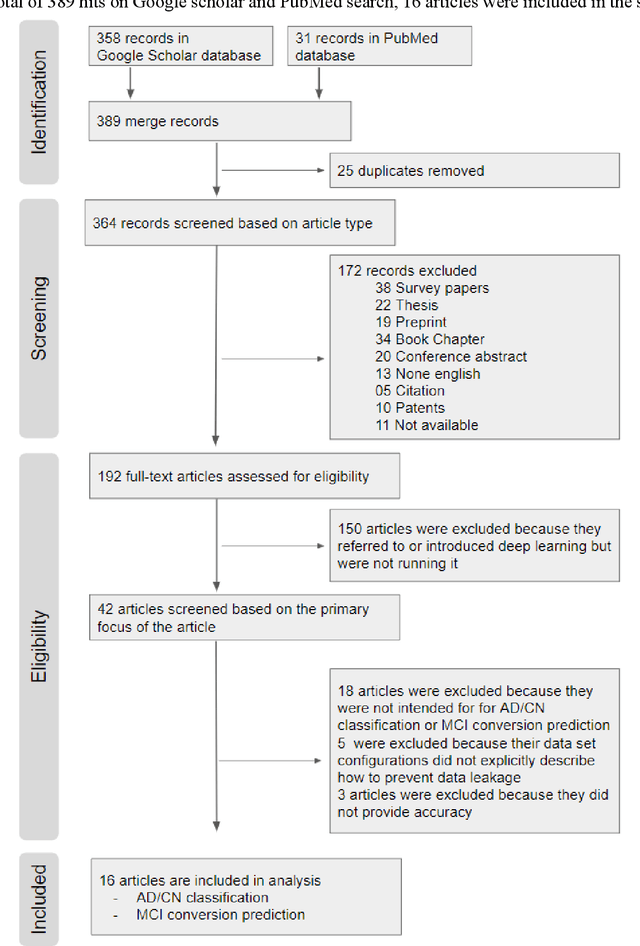
Abstract:The application of deep learning to early detection and automated classification of Alzheimer's disease (AD) has recently gained considerable attention as rapid progress in neuroimaging techniques has generated large-scale multimodal neuroimaging data. Here we systematically reviewed publications, where deep learning approaches and neuroimaging data were used for diagnostic classification of AD. A PubMed and google scholar search was performed to find deep learning papers for AD published between January 2013 and July 2018, which were reviewed, evaluated, and classified by algorithms and neuroimaging types, and findings were summarized. The diagnostic classification of AD using deep learning approaches and neuroimaging data was examined in 16 studies. The approach to combine traditional machine learning for classification and stacked auto-encoder (SAE) for feature selection has produced accuracies of up to 98.8% for AD classification and 83.7% for prediction of conversion from mild cognitive impairment (MCI), a prodromal stage of AD, to AD. Deep learning approaches such as convolutional neural network (CNN) or recurrent neural network (RNN) using neuroimaging data without preprocessing for feature selection have yielded accuracies of up to 96.0% for AD classification and 84.2% for MCI conversion prediction. Furthermore, the best classification performance was obtained when multimodal neuroimaging data as well as fluid biomarkers were integrated. Deep learning approaches without preprocessing neuroimaging data for feature selection, a major bottleneck of traditional machining learning in high-dimensional data, continue to improve their performance and to show great promise in the diagnostic classification of AD using multimodal neuroimaging data.
Evaluation of Protein Structural Models Using Random Forests
Feb 13, 2016



Abstract:Protein structure prediction has been a grand challenge problem in the structure biology over the last few decades. Protein quality assessment plays a very important role in protein structure prediction. In the paper, we propose a new protein quality assessment method which can predict both local and global quality of the protein 3D structural models. Our method uses both multi and single model quality assessment method for global quality assessment, and uses chemical, physical, geo-metrical features, and global quality score for local quality assessment. CASP9 targets are used to generate the features for local quality assessment. We evaluate the performance of our local quality assessment method on CASP10, which is comparable with two stage-of-art QA methods based on the average absolute distance between the real and predicted distance. In addition, we blindly tested our method on CASP11, and the good performance shows that combining single and multiple model quality assessment method could be a good way to improve the accuracy of model quality assessment, and the random forest technique could be used to train a good local quality assessment model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge