Amir Tahmasebi
Supervised Learning in the Presence of Noise: Application in ICD-10 Code Classification
Mar 13, 2021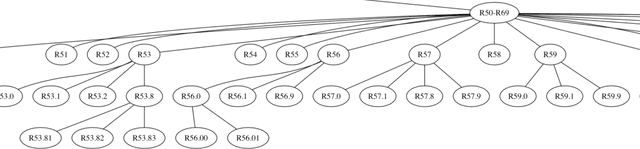
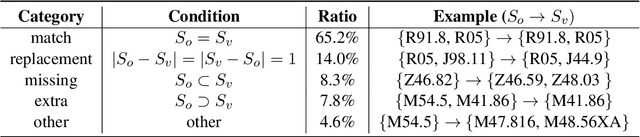
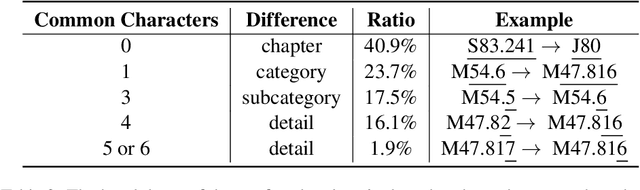
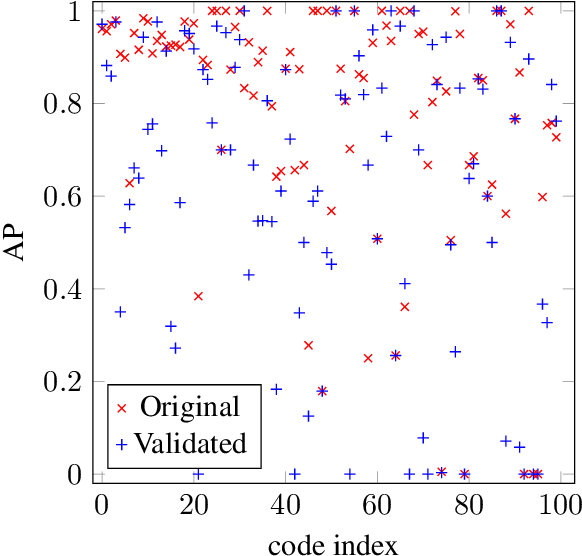
Abstract:ICD coding is the international standard for capturing and reporting health conditions and diagnosis for revenue cycle management in healthcare. Manually assigning ICD codes is prone to human error due to the large code vocabulary and the similarities between codes. Since machine learning based approaches require ground truth training data, the inconsistency among human coders is manifested as noise in labeling, which makes the training and evaluation of ICD classifiers difficult in presence of such noise. This paper investigates the characteristics of such noise in manually-assigned ICD-10 codes and furthermore, proposes a method to train robust ICD-10 classifiers in the presence of labeling noise. Our research concluded that the nature of such noise is systematic. Most of the existing methods for handling label noise assume that the noise is completely random and independent of features or labels, which is not the case for ICD data. Therefore, we develop a new method for training robust classifiers in the presence of systematic noise. We first identify ICD-10 codes that human coders tend to misuse or confuse, based on the codes' locations in the ICD-10 hierarchy, the types of the codes, and baseline classifier's prediction behaviors; we then develop a novel training strategy that accounts for such noise. We compared our method with the baseline that does not handle label noise and the baseline methods that assume random noise, and demonstrated that our proposed method outperforms all baselines when evaluated on expert validated labels.
From Extreme Multi-label to Multi-class: A Hierarchical Approach for Automated ICD-10 Coding Using Phrase-level Attention
Feb 18, 2021



Abstract:Clinical coding is the task of assigning a set of alphanumeric codes, referred to as ICD (International Classification of Diseases), to a medical event based on the context captured in a clinical narrative. The latest version of ICD, ICD-10, includes more than 70,000 codes. As this is a labor-intensive and error-prone task, automatic ICD coding of medical reports using machine learning has gained significant interest in the last decade. Existing literature has modeled this problem as a multi-label task. Nevertheless, such multi-label approach is challenging due to the extremely large label set size. Furthermore, the interpretability of the predictions is essential for the endusers (e.g., healthcare providers and insurance companies). In this paper, we propose a novel approach for automatic ICD coding by reformulating the extreme multi-label problem into a simpler multi-class problem using a hierarchical solution. We made this approach viable through extensive data collection to acquire phrase-level human coder annotations to supervise our models on learning the specific relations between the input text and predicted ICD codes. Our approach employs two independently trained networks, the sentence tagger and the ICD classifier, stacked hierarchically to predict a codeset for a medical report. The sentence tagger identifies focus sentences containing a medical event or concept relevant to an ICD coding. Using a supervised attention mechanism, the ICD classifier then assigns each focus sentence with an ICD code. The proposed approach outperforms strong baselines by large margins of 23% in subset accuracy, 18% in micro-F1, and 15% in instance based F-1. With our proposed approach, interpretability is achieved not through implicitly learned attention scores but by attributing each prediction to a particular sentence and words selected by human coders.
An Ensemble Approach for Automatic Structuring of Radiology Reports
Oct 11, 2020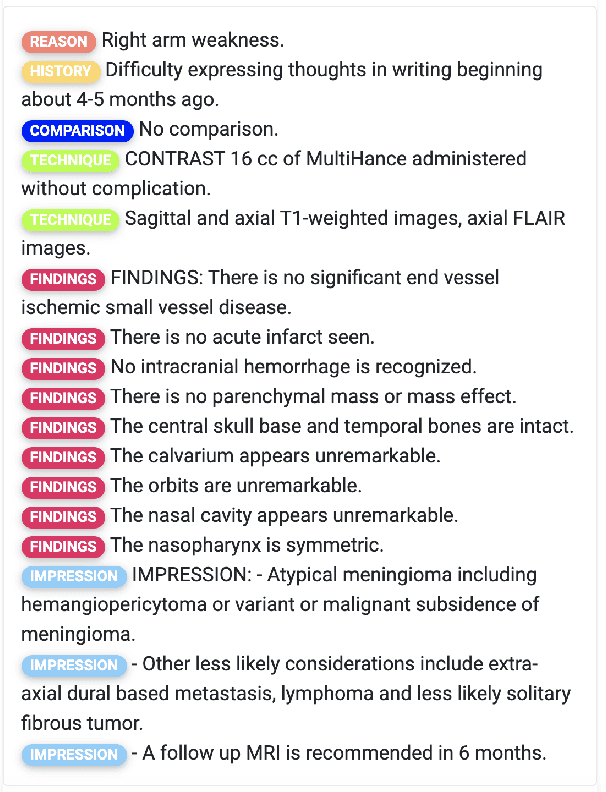
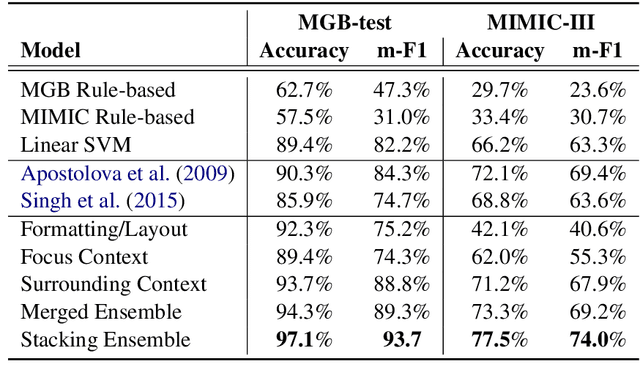
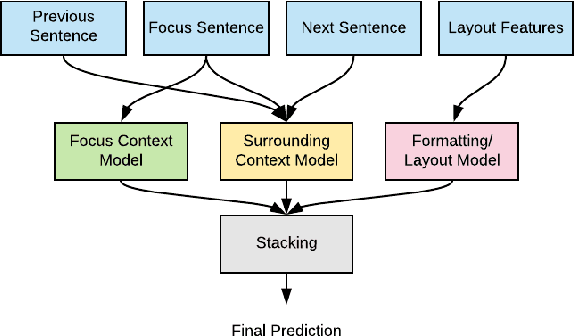
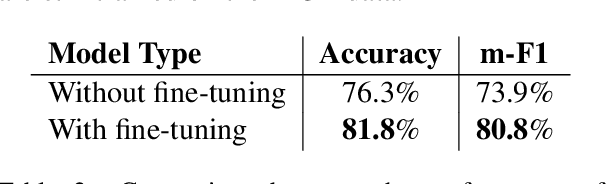
Abstract:Automatic structuring of electronic medical records is of high demand for clinical workflow solutions to facilitate extraction, storage, and querying of patient care information. However, developing a scalable solution is extremely challenging, specifically for radiology reports, as most healthcare institutes use either no template or department/institute specific templates. Moreover, radiologists' reporting style varies from one to another as sentences are telegraphic and do not follow general English grammar rules. We present an ensemble method that consolidates the predictions of three models, capturing various attributes of textual information for automatic labeling of sentences with section labels. These three models are: 1) Focus Sentence model, capturing context of the target sentence; 2) Surrounding Context model, capturing the neighboring context of the target sentence; and finally, 3) Formatting/Layout model, aimed at learning report formatting cues. We utilize Bi-directional LSTMs, followed by sentence encoders, to acquire the context. Furthermore, we define several features that incorporate the structure of reports. We compare our proposed approach against multiple baselines and state-of-the-art approaches on a proprietary dataset as well as 100 manually annotated radiology notes from the MIMIC-III dataset, which we are making publicly available. Our proposed approach significantly outperforms other approaches by achieving 97.1% accuracy.
Clinical Concept Extraction with Contextual Word Embedding
Oct 24, 2018
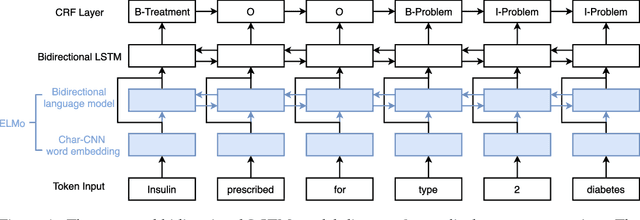


Abstract:Automatic extraction of clinical concepts is an essential step for turning the unstructured data within a clinical note into structured and actionable information. In this work, we propose a clinical concept extraction model for automatic annotation of clinical problems, treatments, and tests in clinical notes utilizing domain-specific contextual word embedding. A contextual word embedding model is first trained on a corpus with a mixture of clinical reports and relevant Wikipedia pages in the clinical domain. Next, a bidirectional LSTM-CRF model is trained for clinical concept extraction using the contextual word embedding model. We tested our proposed model on the I2B2 2010 challenge dataset. Our proposed model achieved the best performance among reported baseline models and outperformed the state-of-the-art models by 3.4% in terms of F1-score.
Towards radiologist-level cancer risk assessment in CT lung screening using deep learning
Apr 05, 2018
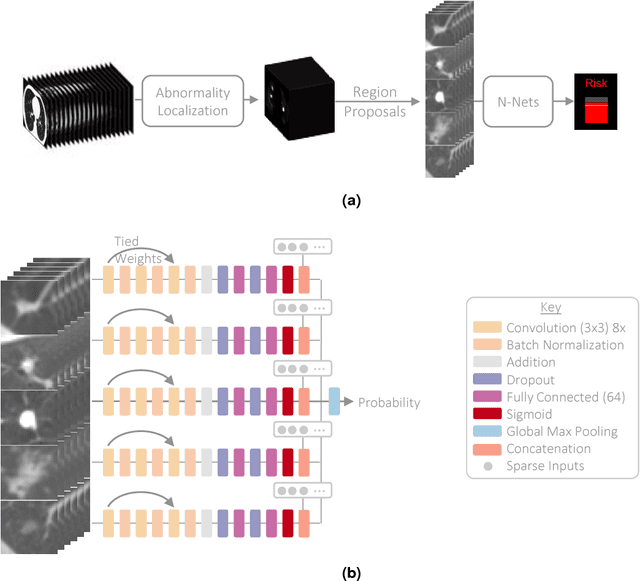
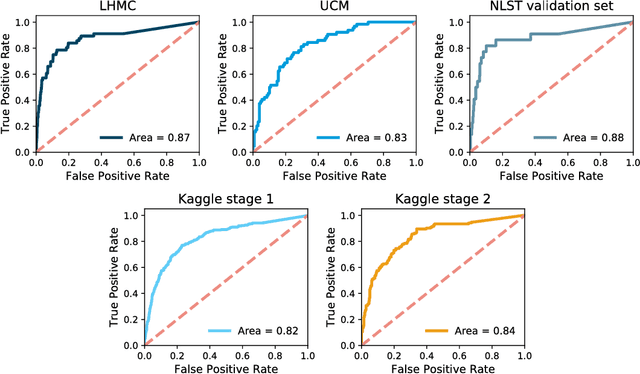
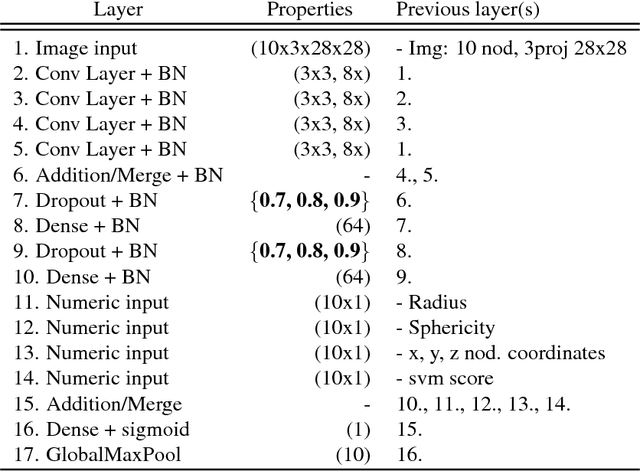
Abstract:Lung cancer is the leading cause of cancer mortality in the US, responsible for more deaths than breast, prostate, colon and pancreas cancer combined. Recently, it has been demonstrated that screening those at high-risk for lung cancer low-dose computed tomography (CT) of the chest can significantly reduce this death rate. The process of evaluating a chest CT scan involves the identification of nodules that are contained within a scan as well as the evaluation of the likelihood that a nodule is malignant based on its imaging characteristics. This has motivated researchers to develop image analysis research tools, such as nodule detectors and nodule classifiers that can assist radiologists to make accurate assessments of the patient cancer risk. In this work, we propose a two-stage framework that can assess the lung cancer risk associated with a low-dose chest CT scan. At the first stage, our framework employs a nodule detector; while in the second stage, we use both the image area around the nodules and nodule features as inputs to a neural network that estimates the malignancy risk of the whole CT scan. The proposed approach: (a) has better performance than the PanCan Risk Model, a widely accepted method for cancer malignancy assessment, achieving around 7% better Area Under Curve score in two independent datasets we have employed; (b) has comparable performance to radiologists in estimating cancer risk at patient level; (c) employs a novel multi-instance weakly-labeled approach to train the deep learning network that requires confirmed cancer diagnosis only at the patient level (not at the nodule level); and (d) employs a large number of lung CT scans (more than 8000) from heterogeneous data sources (NLST, LHMC, and Kaggle competition data) to validate and compare model performance. AUC scores for our model, evaluated against confirmed cancer diagnosis, range between 82% to 90%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge