Alexander Campbell
DBGDGM: Dynamic Brain Graph Deep Generative Model
Jan 26, 2023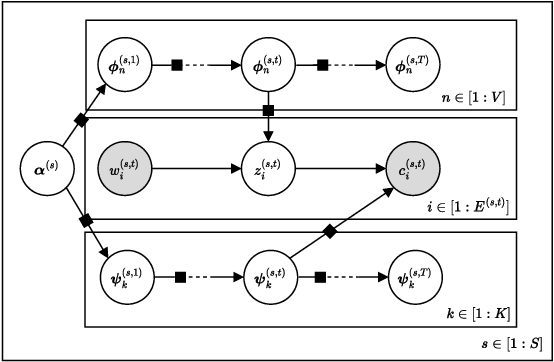
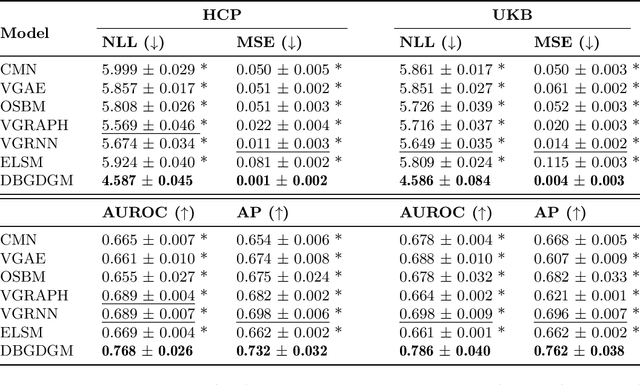
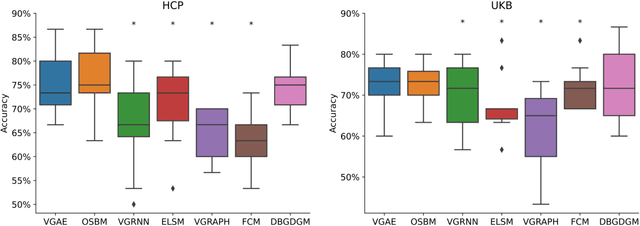
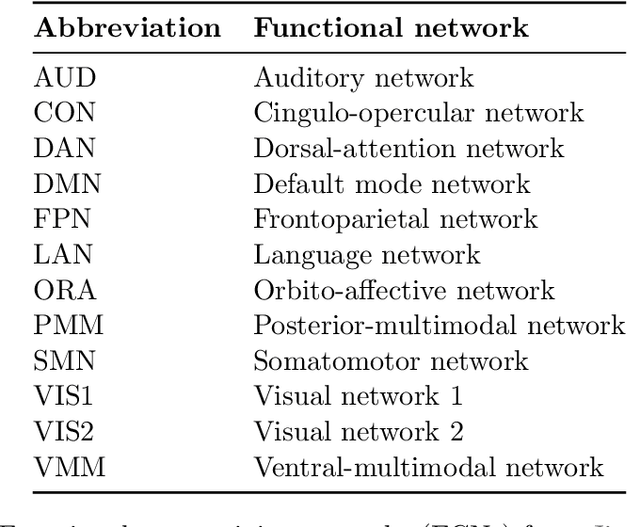
Abstract:Graphs are a natural representation of brain activity derived from functional magnetic imaging (fMRI) data. It is well known that clusters of anatomical brain regions, known as functional connectivity networks (FCNs), encode temporal relationships which can serve as useful biomarkers for understanding brain function and dysfunction. Previous works, however, ignore the temporal dynamics of the brain and focus on static graphs. In this paper, we propose a dynamic brain graph deep generative model (DBGDGM) which simultaneously clusters brain regions into temporally evolving communities and learns dynamic unsupervised node embeddings. Specifically, DBGDGM represents brain graph nodes as embeddings sampled from a distribution over communities that evolve over time. We parameterise this community distribution using neural networks that learn from subject and node embeddings as well as past community assignments. Experiments demonstrate DBGDGM outperforms baselines in graph generation, dynamic link prediction, and is comparable for graph classification. Finally, an analysis of the learnt community distributions reveals overlap with known FCNs reported in neuroscience literature.
DBGSL: Dynamic Brain Graph Structure Learning
Sep 27, 2022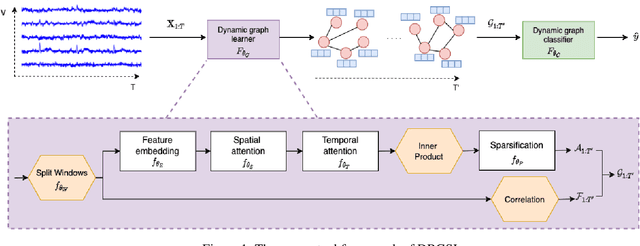
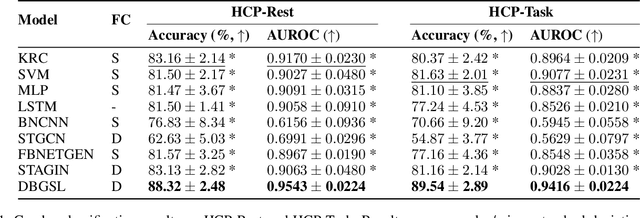
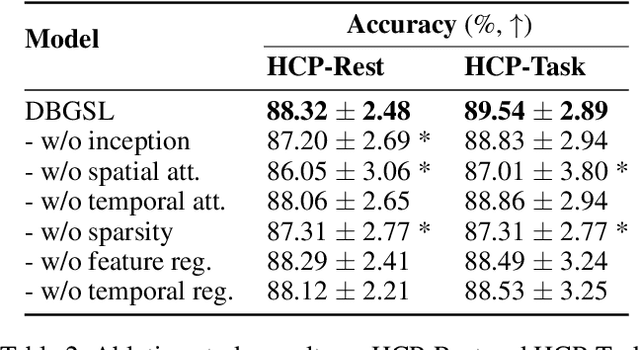
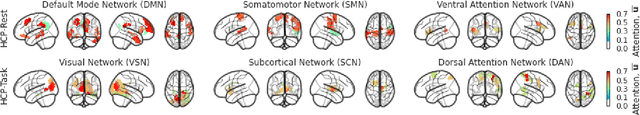
Abstract:Functional connectivity (FC) between regions of the brain is commonly estimated through statistical dependency measures applied to functional magnetic resonance imaging (fMRI) data. The resulting functional connectivity matrix (FCM) is often taken to represent the adjacency matrix of a brain graph. Recently, graph neural networks (GNNs) have been successfully applied to FCMs to learn brain graph representations. A common limitation of existing GNN approaches, however, is that they require the graph adjacency matrix to be known prior to model training. As such, it is implicitly assumed the ground-truth dependency structure of the data is known. Unfortunately, for fMRI this is not the case as the choice of which statistical measure best represents the dependency structure of the data is non-trivial. Also, most GNN applications to fMRI assume FC is static over time, which is at odds with neuroscientific evidence that functional brain networks are time-varying and dynamic. These compounded issues can have a detrimental effect on the capacity of GNNs to learn representations of brain graphs. As a solution, we propose Dynamic Brain Graph Structure Learning (DBGSL), a supervised method for learning the optimal time-varying dependency structure of fMRI data. Specifically, DBGSL learns a dynamic graph from fMRI timeseries via spatial-temporal attention applied to brain region embeddings. The resulting graph is then fed to a spatial-temporal GNN to learn a graph representation for classification. Experiments on large resting-state as well as task fMRI datasets for the task of gender classification demonstrate that DBGSL achieves state-of-the-art performance. Moreover, analysis of the learnt dynamic graphs highlights prediction-related brain regions which align with findings from existing neuroscience literature.
An investigation of pre-upsampling generative modelling and Generative Adversarial Networks in audio super resolution
Sep 30, 2021
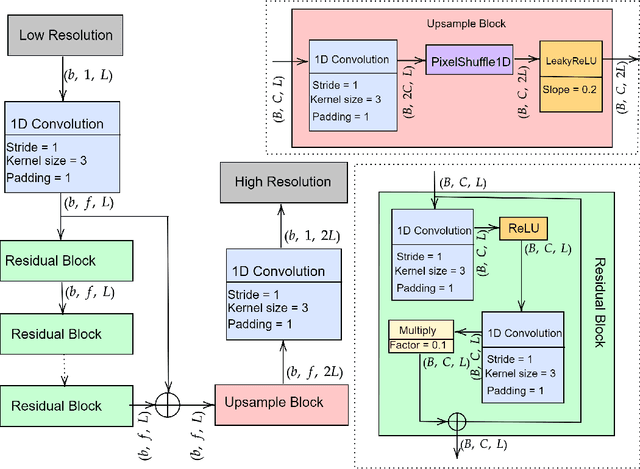
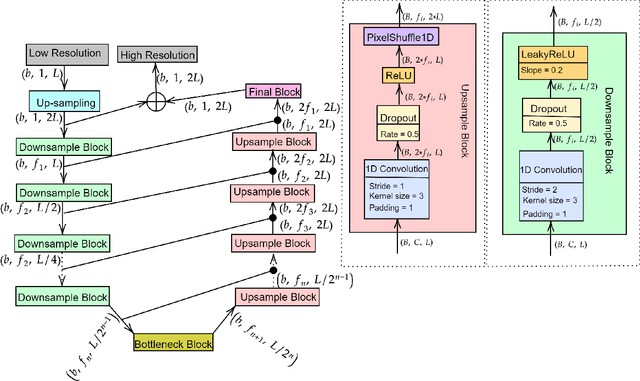
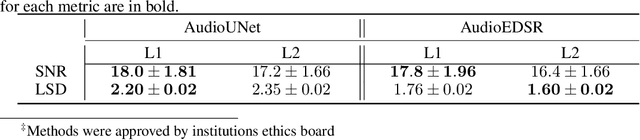
Abstract:There have been several successful deep learning models that perform audio super-resolution. Many of these approaches involve using preprocessed feature extraction which requires a lot of domain-specific signal processing knowledge to implement. Convolutional Neural Networks (CNNs) improved upon this framework by automatically learning filters. An example of a convolutional approach is AudioUNet, which takes inspiration from novel methods of upsampling images. Our paper compares the pre-upsampling AudioUNet to a new generative model that upsamples the signal before using deep learning to transform it into a more believable signal. Based on the EDSR network for image super-resolution, the newly proposed model outperforms UNet with a 20% increase in log spectral distance and a mean opinion score of 4.06 compared to 3.82 for the two times upsampling case. AudioEDSR also has 87% fewer parameters than AudioUNet. How incorporating AudioUNet into a Wasserstein GAN (with gradient penalty) (WGAN-GP) structure can affect training is also explored. Finally the effects artifacting has on the current state of the art is analysed and solutions to this problem are proposed. The methods used in this paper have broad applications to telephony, audio recognition and audio generation tasks.
High Frequency EEG Artifact Detection with Uncertainty via Early Exit Paradigm
Jul 21, 2021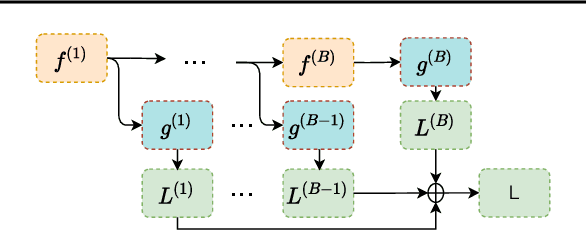
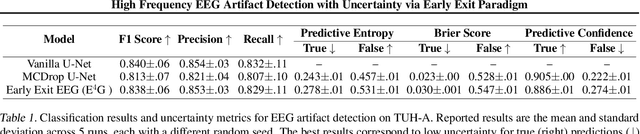
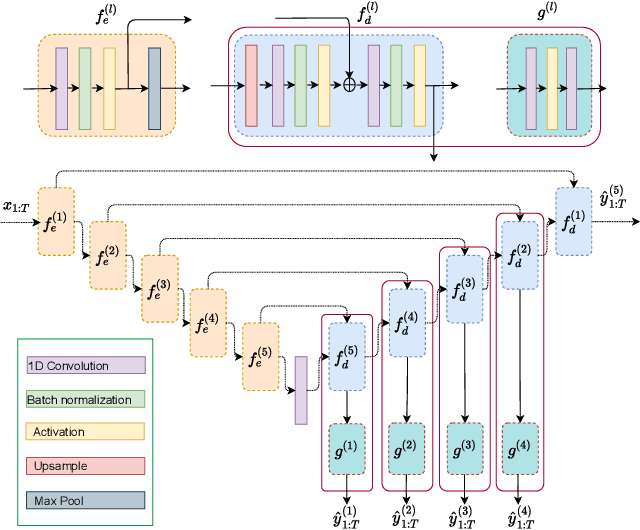
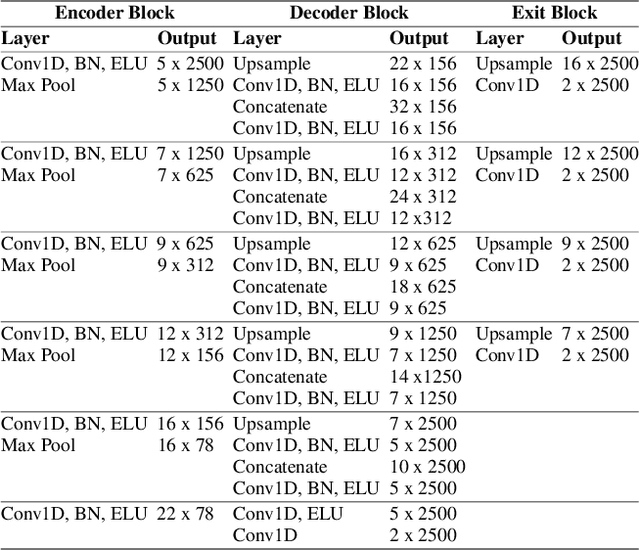
Abstract:Electroencephalography (EEG) is crucial for the monitoring and diagnosis of brain disorders. However, EEG signals suffer from perturbations caused by non-cerebral artifacts limiting their efficacy. Current artifact detection pipelines are resource-hungry and rely heavily on hand-crafted features. Moreover, these pipelines are deterministic in nature, making them unable to capture predictive uncertainty. We propose E4G, a deep learning framework for high frequency EEG artifact detection. Our framework exploits the early exit paradigm, building an implicit ensemble of models capable of capturing uncertainty. We evaluate our approach on the Temple University Hospital EEG Artifact Corpus (v2.0) achieving state-of-the-art classification results. In addition, E4G provides well-calibrated uncertainty metrics comparable to sampling techniques like Monte Carlo dropout in just a single forward pass. E4G opens the door to uncertainty-aware artifact detection supporting clinicians-in-the-loop frameworks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge