Ramon Viñas Torné
An investigation of pre-upsampling generative modelling and Generative Adversarial Networks in audio super resolution
Sep 30, 2021
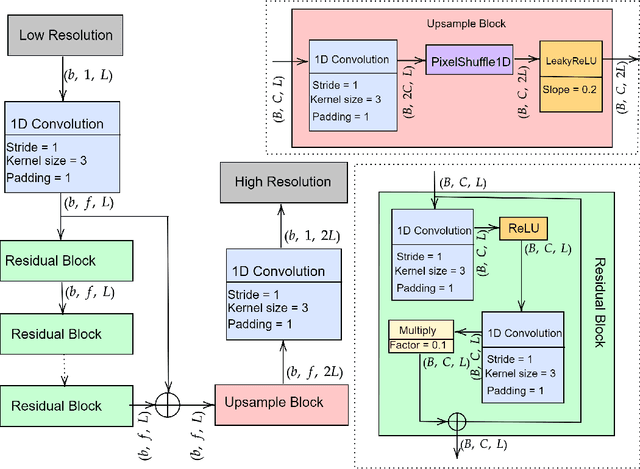
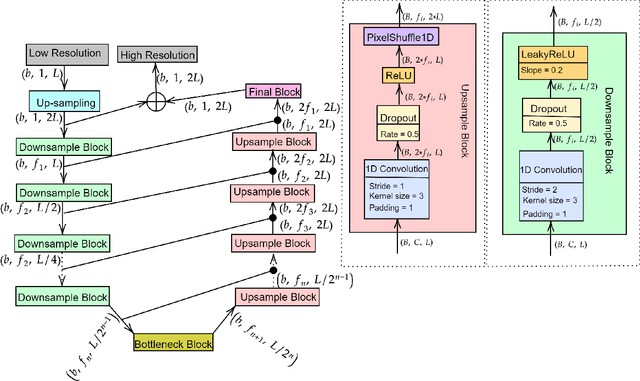
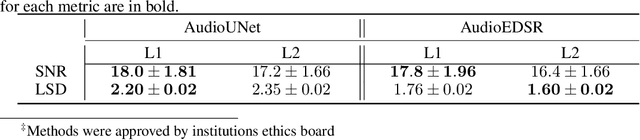
Abstract:There have been several successful deep learning models that perform audio super-resolution. Many of these approaches involve using preprocessed feature extraction which requires a lot of domain-specific signal processing knowledge to implement. Convolutional Neural Networks (CNNs) improved upon this framework by automatically learning filters. An example of a convolutional approach is AudioUNet, which takes inspiration from novel methods of upsampling images. Our paper compares the pre-upsampling AudioUNet to a new generative model that upsamples the signal before using deep learning to transform it into a more believable signal. Based on the EDSR network for image super-resolution, the newly proposed model outperforms UNet with a 20% increase in log spectral distance and a mean opinion score of 4.06 compared to 3.82 for the two times upsampling case. AudioEDSR also has 87% fewer parameters than AudioUNet. How incorporating AudioUNet into a Wasserstein GAN (with gradient penalty) (WGAN-GP) structure can affect training is also explored. Finally the effects artifacting has on the current state of the art is analysed and solutions to this problem are proposed. The methods used in this paper have broad applications to telephony, audio recognition and audio generation tasks.
Graph Representation Learning on Tissue-Specific Multi-Omics
Jul 25, 2021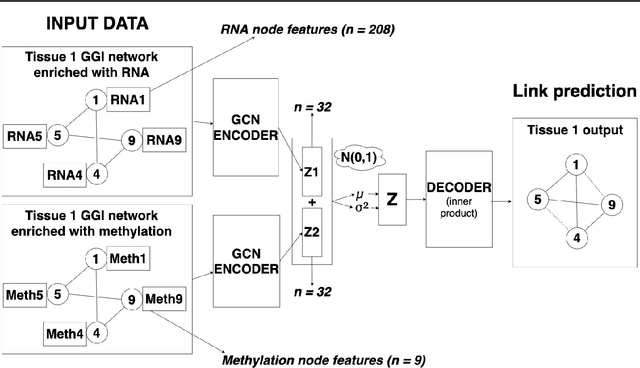
Abstract:Combining different modalities of data from human tissues has been critical in advancing biomedical research and personalised medical care. In this study, we leverage a graph embedding model (i.e VGAE) to perform link prediction on tissue-specific Gene-Gene Interaction (GGI) networks. Through ablation experiments, we prove that the combination of multiple biological modalities (i.e multi-omics) leads to powerful embeddings and better link prediction performances. Our evaluation shows that the integration of gene methylation profiles and RNA-sequencing data significantly improves the link prediction performance. Overall, the combination of RNA-sequencing and gene methylation data leads to a link prediction accuracy of 71% on GGI networks. By harnessing graph representation learning on multi-omics data, our work brings novel insights to the current literature on multi-omics integration in bioinformatics.
Graph representation forecasting of patient's medical conditions: towards a digital twin
Sep 17, 2020
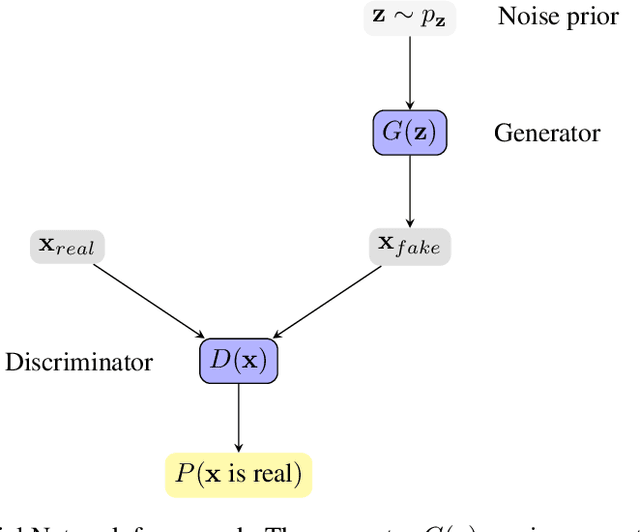
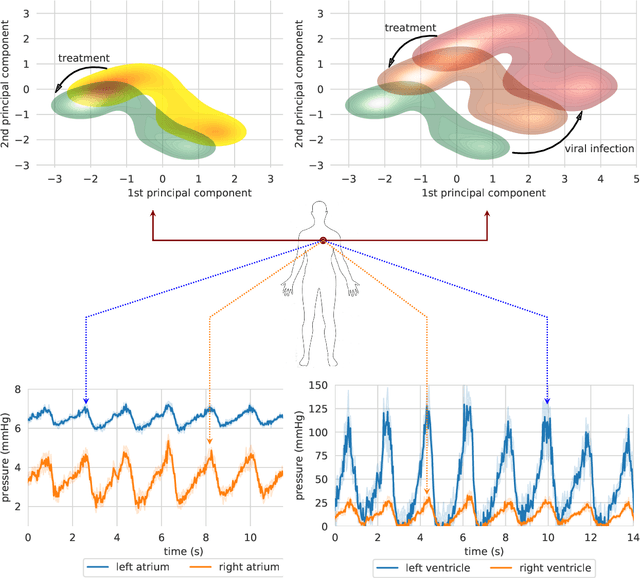
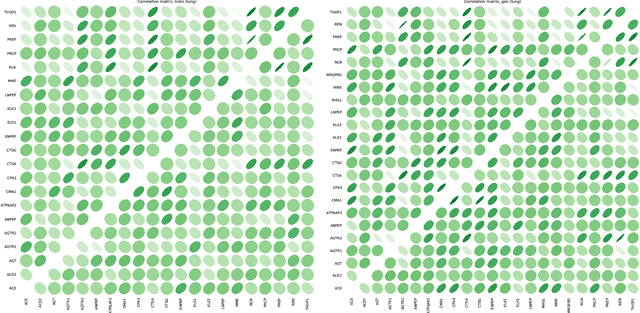
Abstract:Objective: Modern medicine needs to shift from a wait and react, curative discipline to a preventative, interdisciplinary science aiming at providing personalised, systemic and precise treatment plans to patients. The aim of this work is to present how the integration of machine learning approaches with mechanistic computational modelling could yield a reliable infrastructure to run probabilistic simulations where the entire organism is considered as a whole. Methods: We propose a general framework that composes advanced AI approaches and integrates mathematical modelling in order to provide a panoramic view over current and future physiological conditions. The proposed architecture is based on a graph neural network (GNNs) forecasting clinically relevant endpoints (such as blood pressure) and a generative adversarial network (GANs) providing a proof of concept of transcriptomic integrability. Results: We show the results of the investigation of pathological effects of overexpression of ACE2 across different signalling pathways in multiple tissues on cardiovascular functions. We provide a proof of concept of integrating a large set of composable clinical models using molecular data to drive local and global clinical parameters and derive future trajectories representing the evolution of the physiological state of the patient. Significance: We argue that the graph representation of a computational patient has potential to solve important technological challenges in integrating multiscale computational modelling with AI. We believe that this work represents a step forward towards a healthcare digital twin.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge