Alcides Fonseca
Call for Action: towards the next generation of symbolic regression benchmark
May 06, 2025


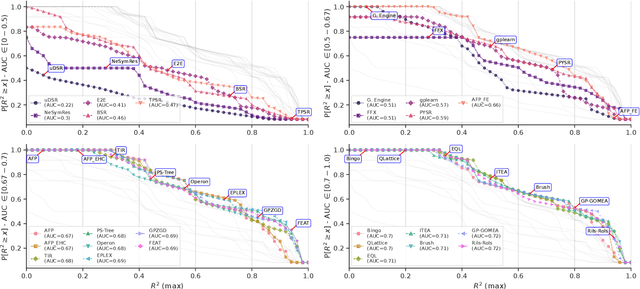
Abstract:Symbolic Regression (SR) is a powerful technique for discovering interpretable mathematical expressions. However, benchmarking SR methods remains challenging due to the diversity of algorithms, datasets, and evaluation criteria. In this work, we present an updated version of SRBench. Our benchmark expands the previous one by nearly doubling the number of evaluated methods, refining evaluation metrics, and using improved visualizations of the results to understand the performances. Additionally, we analyze trade-offs between model complexity, accuracy, and energy consumption. Our results show that no single algorithm dominates across all datasets. We propose a call for action from SR community in maintaining and evolving SRBench as a living benchmark that reflects the state-of-the-art in symbolic regression, by standardizing hyperparameter tuning, execution constraints, and computational resource allocation. We also propose deprecation criteria to maintain the benchmark's relevance and discuss best practices for improving SR algorithms, such as adaptive hyperparameter tuning and energy-efficient implementations.
Understanding Misconfigurations in ROS: An Empirical Study and Current Approaches
Jul 27, 2024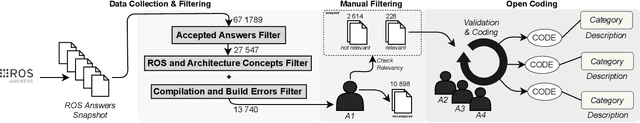
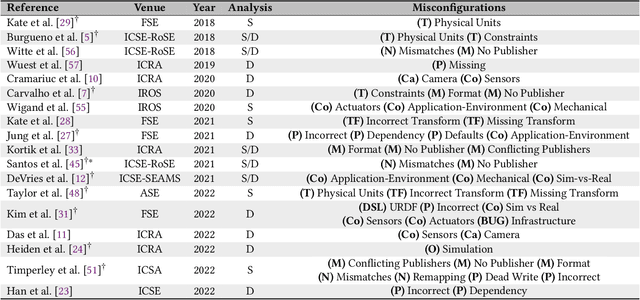
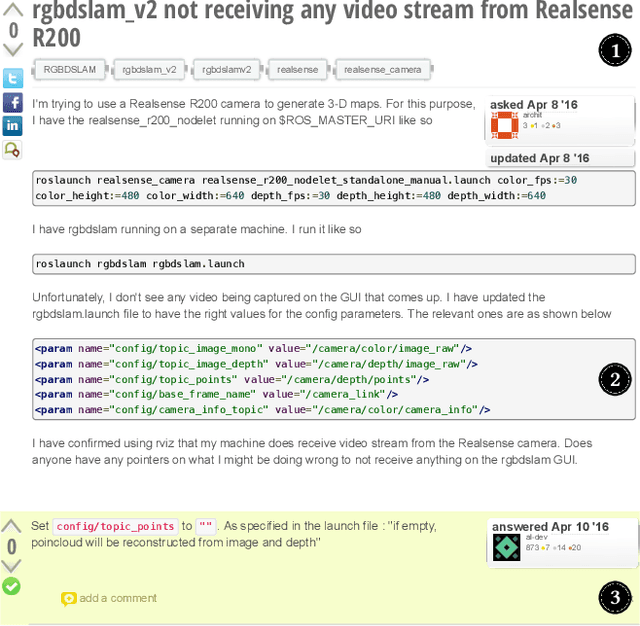
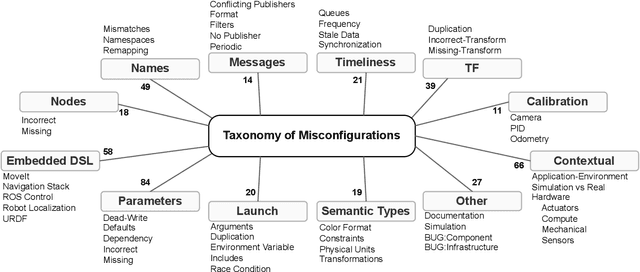
Abstract:The Robot Operating System (ROS) is a popular framework and ecosystem that allows developers to build robot software systems from reusable, off-the-shelf components. Systems are often built by customizing and connecting components via configuration files. While reusable components theoretically allow rapid prototyping, ensuring proper configuration and connection is challenging, as evidenced by numerous questions on developer forums. Developers must abide to the often unchecked and unstated assumptions of individual components. Failure to do so can result in misconfigurations that are only discovered during field deployment, at which point errors may lead to unpredictable and dangerous behavior. Despite misconfigurations having been studied in the broader context of software engineering, robotics software (and ROS in particular) poses domain-specific challenges with potentially disastrous consequences. To understand and improve the reliability of ROS projects, it is critical to identify the types of misconfigurations faced by developers. To that end, we perform a study of ROS Answers, a Q&A platform, to identify and categorize misconfigurations that occur during ROS development. We then conduct a literature review to assess the coverage of these misconfigurations by existing detection techniques. In total, we find 12 high-level categories and 50 sub-categories of misconfigurations. Of these categories, 27 are not covered by existing techniques. To conclude, we discuss how to tackle those misconfigurations in future work.
Semantically Rich Local Dataset Generation for Explainable AI in Genomics
Jul 03, 2024



Abstract:Black box deep learning models trained on genomic sequences excel at predicting the outcomes of different gene regulatory mechanisms. Therefore, interpreting these models may provide novel insights into the underlying biology, supporting downstream biomedical applications. Due to their complexity, interpretable surrogate models can only be built for local explanations (e.g., a single instance). However, accomplishing this requires generating a dataset in the neighborhood of the input, which must maintain syntactic similarity to the original data while introducing semantic variability in the model's predictions. This task is challenging due to the complex sequence-to-function relationship of DNA. We propose using Genetic Programming to generate datasets by evolving perturbations in sequences that contribute to their semantic diversity. Our custom, domain-guided individual representation effectively constrains syntactic similarity, and we provide two alternative fitness functions that promote diversity with no computational effort. Applied to the RNA splicing domain, our approach quickly achieves good diversity and significantly outperforms a random baseline in exploring the search space, as shown by our proof-of-concept, short RNA sequence. Furthermore, we assess its generalizability and demonstrate scalability to larger sequences, resulting in a $\approx$30\% improvement over the baseline.
Data types as a more ergonomic frontend for Grammar-Guided Genetic Programming
Oct 10, 2022

Abstract:Genetic Programming (GP) is an heuristic method that can be applied to many Machine Learning, Optimization and Engineering problems. In particular, it has been widely used in Software Engineering for Test-case generation, Program Synthesis and Improvement of Software (GI). Grammar-Guided Genetic Programming (GGGP) approaches allow the user to refine the domain of valid program solutions. Backus Normal Form is the most popular interface for describing Context-Free Grammars (CFG) for GGGP. BNF and its derivatives have the disadvantage of interleaving the grammar language and the target language of the program. We propose to embed the grammar as an internal Domain-Specific Language in the host language of the framework. This approach has the same expressive power as BNF and EBNF while using the host language type-system to take advantage of all the existing tooling: linters, formatters, type-checkers, autocomplete, and legacy code support. These tools have a practical utility in designing software in general, and GP systems in particular. We also present Meta-Handlers, user-defined overrides of the tree-generation system. This technique extends our object-oriented encoding with more practicability and expressive power than existing CFG approaches, achieving the same expressive power of Attribute Grammars, but without the grammar vs target language duality. Furthermore, we evidence that this approach is feasible, showing an example Python implementation as proof. We also compare our approach against textual BNF-representations w.r.t. expressive power and ergonomics. These advantages do not come at the cost of performance, as shown by our empirical evaluation on 5 benchmarks of our example implementation against PonyGE2. We conclude that our approach has better ergonomics with the same expressive power and performance of textual BNF-based grammar encodings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge