Albert Cardona
Learning to rank via combining representations
May 20, 2020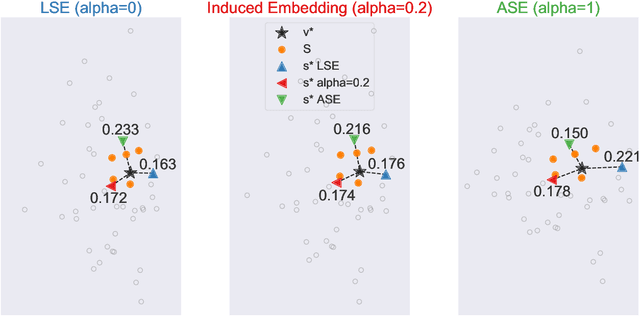
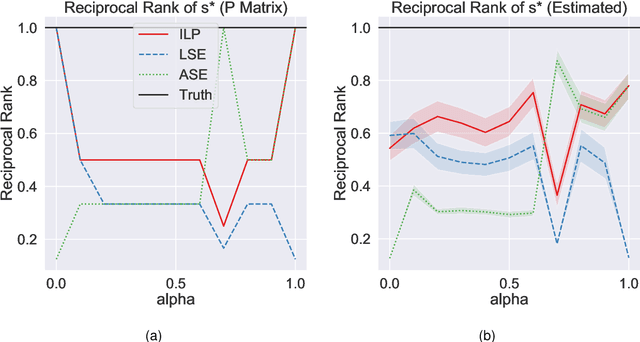
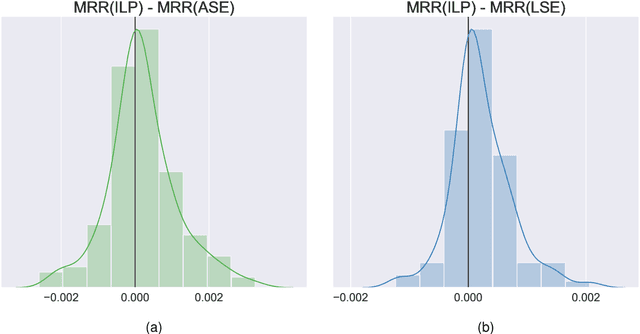
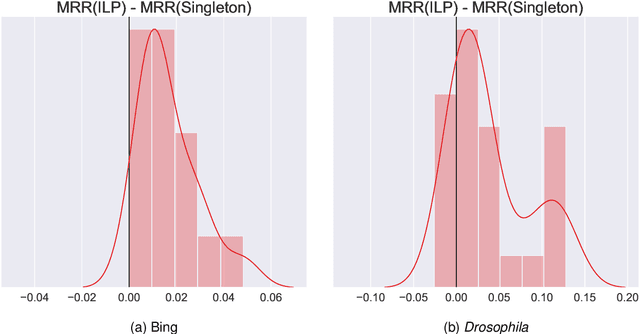
Abstract:Learning to rank -- producing a ranked list of items specific to a query and with respect to a set of supervisory items -- is a problem of general interest. The setting we consider is one in which no analytic description of what constitutes a good ranking is available. Instead, we have a collection of representations and supervisory information consisting of a (target item, interesting items set) pair. We demonstrate -- analytically, in simulation, and in real data examples -- that learning to rank via combining representations using an integer linear program is effective when the supervision is as light as "these few items are similar to your item of interest." While this nomination task is of general interest, for specificity we present our methodology from the perspective of vertex nomination in graphs. The methodology described herein is model agnostic.
Semiparametric spectral modeling of the Drosophila connectome
May 09, 2017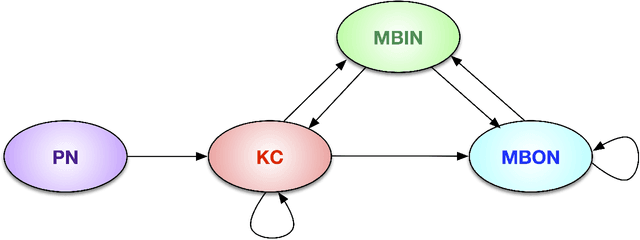
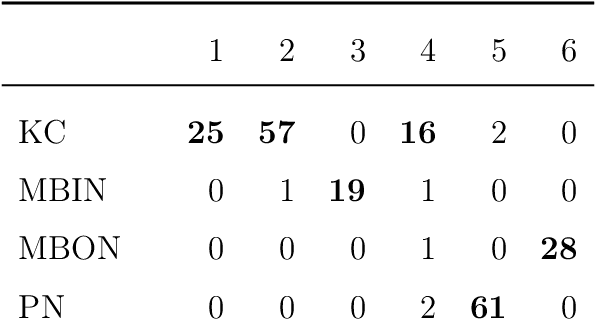
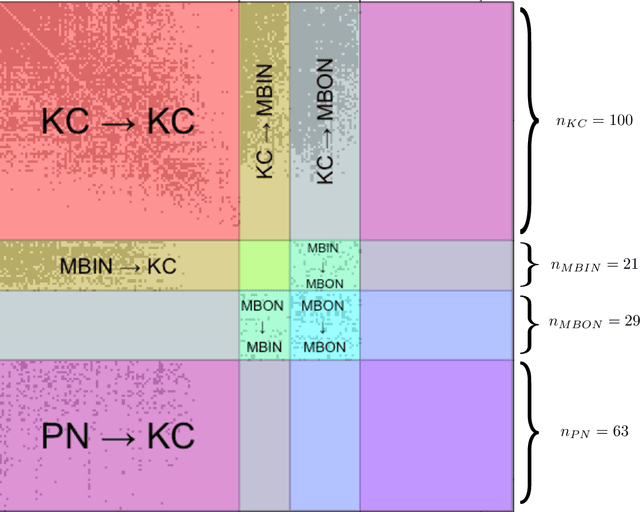
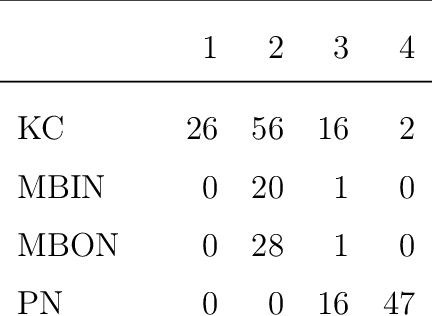
Abstract:We present semiparametric spectral modeling of the complete larval Drosophila mushroom body connectome. Motivated by a thorough exploratory data analysis of the network via Gaussian mixture modeling (GMM) in the adjacency spectral embedding (ASE) representation space, we introduce the latent structure model (LSM) for network modeling and inference. LSM is a generalization of the stochastic block model (SBM) and a special case of the random dot product graph (RDPG) latent position model, and is amenable to semiparametric GMM in the ASE representation space. The resulting connectome code derived via semiparametric GMM composed with ASE captures latent connectome structure and elucidates biologically relevant neuronal properties.
TED: A Tolerant Edit Distance for Segmentation Evaluation
Feb 01, 2016


Abstract:In this paper, we present a novel error measure to compare a segmentation against ground truth. This measure, which we call Tolerant Edit Distance (TED), is motivated by two observations: (1) Some errors, like small boundary shifts, are tolerable in practice. Which errors are tolerable is application dependent and should be a parameter of the measure. (2) Non-tolerable errors have to be corrected manually. The time needed to do so should be reflected by the error measure. Using integer linear programming, the TED finds the minimal weighted sum of split and merge errors exceeding a given tolerance criterion, and thus provides a time-to-fix estimate. In contrast to commonly used measures like Rand index or variation of information, the TED (1) does not count small, but tolerable, differences, (2) provides intuitive numbers, (3) gives a time-to-fix estimate, and (4) can localize and classify the type of errors. By supporting both isotropic and anisotropic volumes and having a flexible tolerance criterion, the TED can be adapted to different requirements. On example segmentations for 3D neuron segmentation, we demonstrate that the TED is capable of counting topological errors, while ignoring small boundary shifts.
Multi-Hypothesis CRF-Segmentation of Neural Tissue in Anisotropic EM Volumes
Sep 18, 2011



Abstract:We present an approach for the joint segmentation and grouping of similar components in anisotropic 3D image data and use it to segment neural tissue in serial sections electron microscopy (EM) images. We first construct a nested set of neuron segmentation hypotheses for each slice. A conditional random field (CRF) then allows us to evaluate both the compatibility of a specific segmentation and a specific inter-slice assignment of neuron candidates with the underlying observations. The model is solved optimally for an entire image stack simultaneously using integer linear programming (ILP), which yields the maximum a posteriori solution in amortized linear time in the number of slices. We evaluate the performance of our approach on an annotated sample of the Drosophila larva neuropil and show that the consideration of different segmentation hypotheses in each slice leads to a significant improvement in the segmentation and assignment accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge