Adam Cross
Integration of Large Language Models and Traditional Deep Learning for Social Determinants of Health Prediction
May 06, 2025Abstract:Social Determinants of Health (SDoH) are economic, social and personal circumstances that affect or influence an individual's health status. SDoHs have shown to be correlated to wellness outcomes, and therefore, are useful to physicians in diagnosing diseases and in decision-making. In this work, we automatically extract SDoHs from clinical text using traditional deep learning and Large Language Models (LLMs) to find the advantages and disadvantages of each on an existing publicly available dataset. Our models outperform a previous reference point on a multilabel SDoH classification by 10 points, and we present a method and model to drastically speed up classification (12X execution time) by eliminating expensive LLM processing. The method we present combines a more nimble and efficient solution that leverages the power of the LLM for precision and traditional deep learning methods for efficiency. We also show highly performant results on a dataset supplemented with synthetic data and several traditional deep learning models that outperform LLMs. Our models and methods offer the next iteration of automatic prediction of SDoHs that impact at-risk patients.
Leveraging LLMs for Predicting Unknown Diagnoses from Clinical Notes
Mar 28, 2025Abstract:Electronic Health Records (EHRs) often lack explicit links between medications and diagnoses, making clinical decision-making and research more difficult. Even when links exist, diagnosis lists may be incomplete, especially during early patient visits. Discharge summaries tend to provide more complete information, which can help infer accurate diagnoses, especially with the help of large language models (LLMs). This study investigates whether LLMs can predict implicitly mentioned diagnoses from clinical notes and link them to corresponding medications. We address two research questions: (1) Does majority voting across diverse LLM configurations outperform the best single configuration in diagnosis prediction? (2) How sensitive is majority voting accuracy to LLM hyperparameters such as temperature, top-p, and summary length? To evaluate, we created a new dataset of 240 expert-annotated medication-diagnosis pairs from 20 MIMIC-IV notes. Using GPT-3.5 Turbo, we ran 18 prompting configurations across short and long summary lengths, generating 8568 test cases. Results show that majority voting achieved 75 percent accuracy, outperforming the best single configuration at 66 percent. No single hyperparameter setting dominated, but combining deterministic, balanced, and exploratory strategies improved performance. Shorter summaries generally led to higher accuracy.In conclusion, ensemble-style majority voting with diverse LLM configurations improves diagnosis prediction in EHRs and offers a promising method to link medications and diagnoses in clinical texts.
AutoRD: An Automatic and End-to-End System for Rare Disease Knowledge Graph Construction Based on Ontologies-enhanced Large Language Models
Mar 01, 2024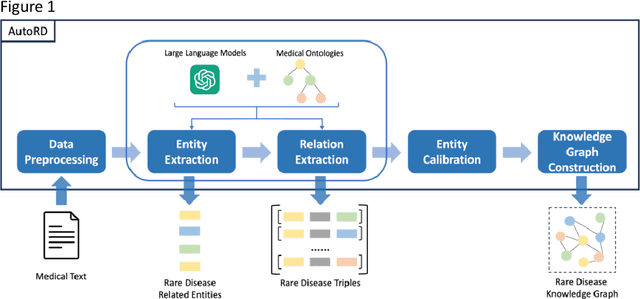


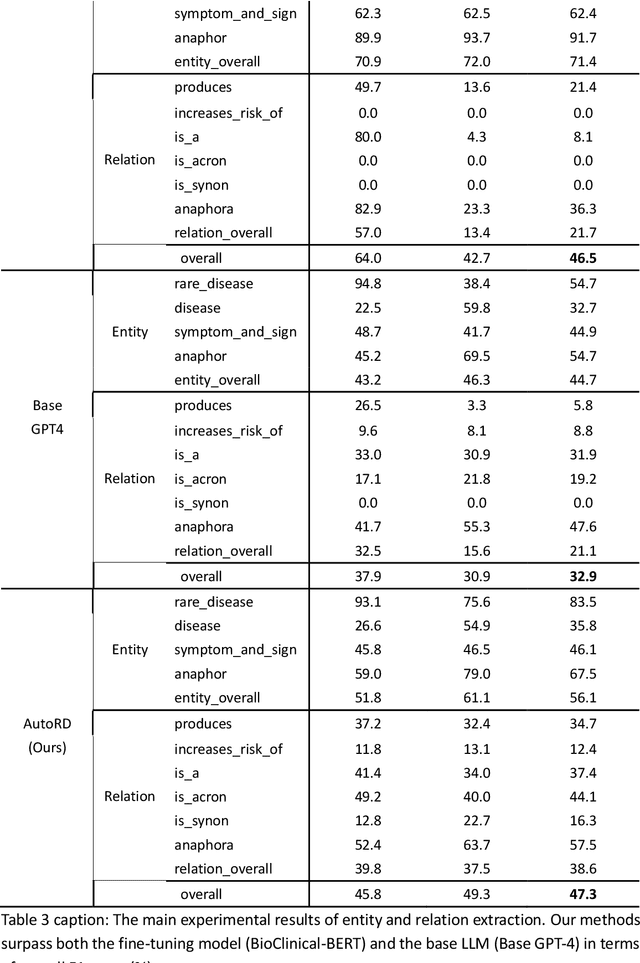
Abstract:Objectives: Our objective is to create an end-to-end system called AutoRD, which automates extracting information from clinical text about rare diseases. We have conducted various tests to evaluate the performance of AutoRD and highlighted its strengths and limitations in this paper. Materials and Methods: Our system, AutoRD, is a software pipeline involving data preprocessing, entity extraction, relation extraction, entity calibration, and knowledge graph construction. We implement this using large language models and medical knowledge graphs developed from open-source medical ontologies. We quantitatively evaluate our system on entity extraction, relation extraction, and the performance of knowledge graph construction. Results: AutoRD achieves an overall F1 score of 47.3%, a 14.4% improvement compared to the base LLM. In detail, AutoRD achieves an overall entity extraction F1 score of 56.1% (rare_disease: 83.5%, disease: 35.8%, symptom_and_sign: 46.1%, anaphor: 67.5%) and an overall relation extraction F1 score of 38.6% (produces: 34.7%, increases_risk_of: 12.4%, is_a: 37.4%, is_acronym: 44.1%, is_synonym: 16.3%, anaphora: 57.5%). Our qualitative experiment also demonstrates that the performance in constructing the knowledge graph is commendable. Discussion: AutoRD demonstrates the potential of LLM applications in rare disease detection. This improvement is attributed to several design, including the integration of ontologies-enhanced LLMs. Conclusion: AutoRD is an automated end-to-end system for extracting rare disease information from text to build knowledge graphs. It uses ontologies-enhanced LLMs for a robust medical knowledge base. The superior performance of AutoRD is validated by experimental evaluations, demonstrating the potential of LLMs in healthcare.
GraphCare: Enhancing Healthcare Predictions with Open-World Personalized Knowledge Graphs
May 22, 2023Abstract:Clinical predictive models often rely on patients electronic health records (EHR), but integrating medical knowledge to enhance predictions and decision-making is challenging. This is because personalized predictions require personalized knowledge graphs (KGs), which are difficult to generate from patient EHR data. To address this, we propose GraphCare, an open-world framework that leverages external KGs to improve EHR-based predictions. Our method extracts knowledge from large language models (LLMs) and external biomedical KGs to generate patient-specific KGs, which are then used to train our proposed Bi-attention AugmenTed BAT graph neural network GNN for healthcare predictions. We evaluate GraphCare on two public datasets: MIMIC-III and MIMIC-IV. Our method outperforms baseline models in four vital healthcare prediction tasks: mortality, readmission, length-of-stay, and drug recommendation, improving AUROC on MIMIC-III by average margins of 10.4%, 3.8%, 2.0%, and 1.5%, respectively. Notably, GraphCare demonstrates a substantial edge in scenarios with limited data availability. Our findings highlight the potential of using external KGs in healthcare prediction tasks and demonstrate the promise of GraphCare in generating personalized KGs for promoting personalized medicine.
MedML: Fusing Medical Knowledge and Machine Learning Models for Early Pediatric COVID-19 Hospitalization and Severity Prediction
Jul 25, 2022


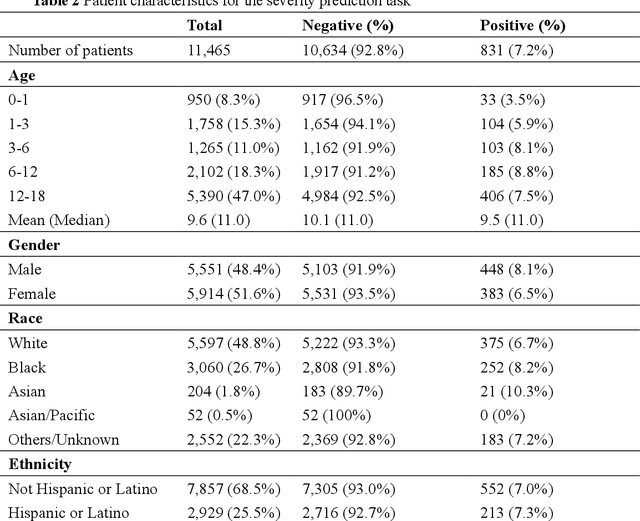
Abstract:The COVID-19 pandemic has caused devastating economic and social disruption, straining the resources of healthcare institutions worldwide. This has led to a nationwide call for models to predict hospitalization and severe illness in patients with COVID-19 to inform distribution of limited healthcare resources. We respond to one of these calls specific to the pediatric population. To address this challenge, we study two prediction tasks for the pediatric population using electronic health records: 1) predicting which children are more likely to be hospitalized, and 2) among hospitalized children, which individuals are more likely to develop severe symptoms. We respond to the national Pediatric COVID-19 data challenge with a novel machine learning model, MedML. MedML extracts the most predictive features based on medical knowledge and propensity scores from over 6 million medical concepts and incorporates the inter-feature relationships between heterogeneous medical features via graph neural networks (GNN). We evaluate MedML across 143,605 patients for the hospitalization prediction task and 11,465 patients for the severity prediction task using data from the National Cohort Collaborative (N3C) dataset. We also report detailed group-level and individual-level feature importance analyses to evaluate the model interpretability. MedML achieves up to a 7% higher AUROC score and up to a 14% higher AUPRC score compared to the best baseline machine learning models and performs well across all nine national geographic regions and over all three-month spans since the start of the pandemic. Our cross-disciplinary research team has developed a method of incorporating clinical domain knowledge as the framework for a new type of machine learning model that is more predictive and explainable than current state-of-the-art data-driven feature selection methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge